| Full name: cryptochrome circadian regulator 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11p11.2 | ||
| Entrez ID: 1408 | HGNC ID: HGNC:2385 | Ensembl Gene: ENSG00000121671 | OMIM ID: 603732 |
| Drug and gene relationship at DGIdb | |||
CRY2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04710 | Circadian rhythm |
Expression of CRY2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRY2 | 1408 | 212695_at | -0.4990 | 0.4079 | |
| GSE20347 | CRY2 | 1408 | 212695_at | -0.3154 | 0.0012 | |
| GSE23400 | CRY2 | 1408 | 212695_at | -0.2995 | 0.0000 | |
| GSE26886 | CRY2 | 1408 | 212695_at | -0.3193 | 0.0429 | |
| GSE29001 | CRY2 | 1408 | 212695_at | -0.3607 | 0.0346 | |
| GSE38129 | CRY2 | 1408 | 212695_at | -0.6561 | 0.0000 | |
| GSE45670 | CRY2 | 1408 | 212695_at | -0.3272 | 0.0206 | |
| GSE53622 | CRY2 | 1408 | 48884 | -0.8257 | 0.0000 | |
| GSE53624 | CRY2 | 1408 | 48884 | -0.6904 | 0.0000 | |
| GSE63941 | CRY2 | 1408 | 212695_at | -0.4998 | 0.0326 | |
| GSE77861 | CRY2 | 1408 | 212695_at | -0.1162 | 0.2690 | |
| GSE97050 | CRY2 | 1408 | A_23_P127394 | -0.4976 | 0.2379 | |
| SRP007169 | CRY2 | 1408 | RNAseq | -1.1341 | 0.0029 | |
| SRP008496 | CRY2 | 1408 | RNAseq | -0.7160 | 0.0289 | |
| SRP064894 | CRY2 | 1408 | RNAseq | -0.5827 | 0.0025 | |
| SRP133303 | CRY2 | 1408 | RNAseq | -0.1772 | 0.5830 | |
| SRP159526 | CRY2 | 1408 | RNAseq | -0.5411 | 0.1198 | |
| SRP193095 | CRY2 | 1408 | RNAseq | -0.5859 | 0.0000 | |
| SRP219564 | CRY2 | 1408 | RNAseq | -0.5078 | 0.4393 | |
| TCGA | CRY2 | 1408 | RNAseq | -0.5645 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
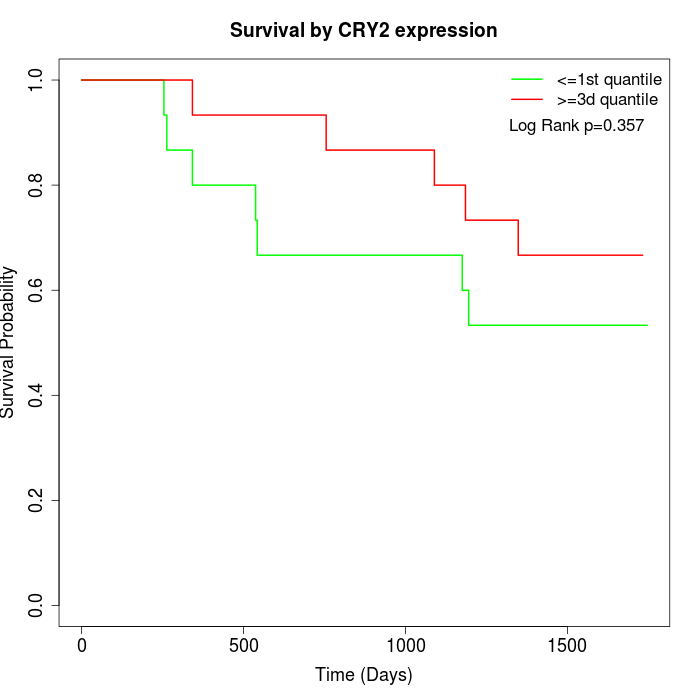
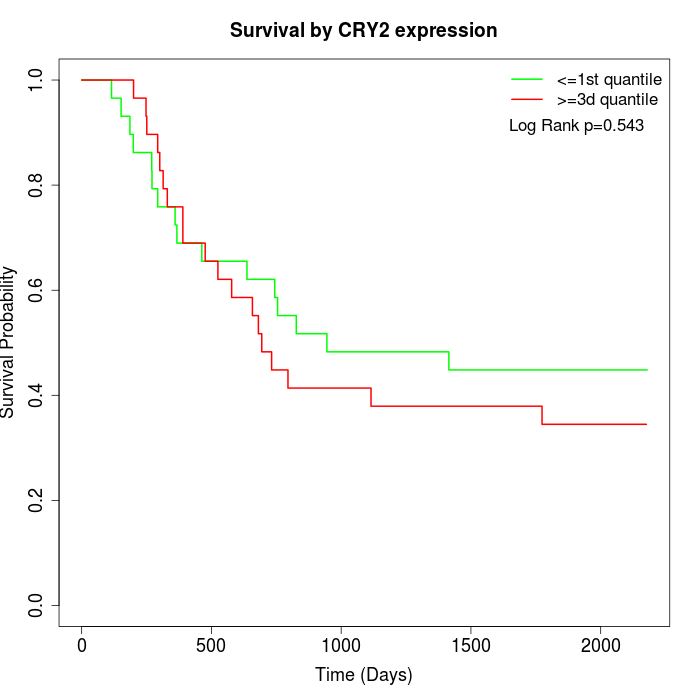
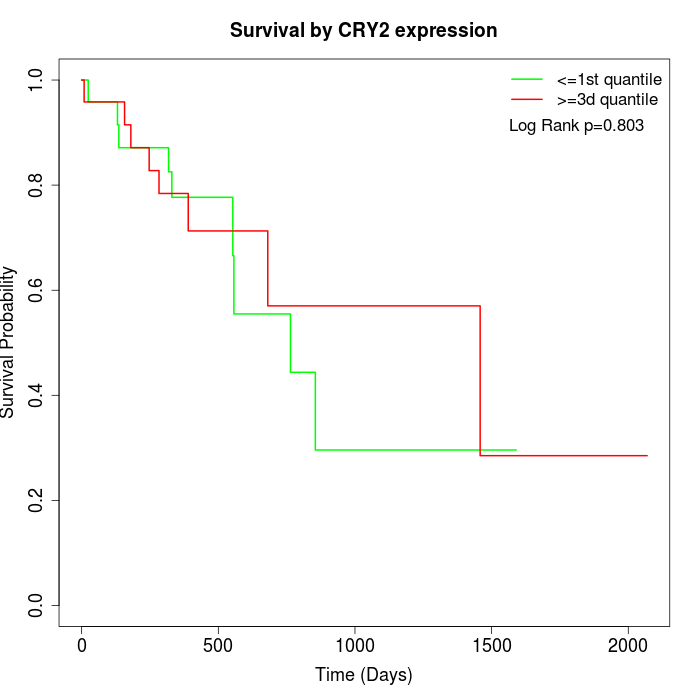
Survival by CRY2 expression:
Note: Click image to view full size file.
Copy number change of CRY2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRY2 | 1408 | 3 | 7 | 20 | |
| GSE20123 | CRY2 | 1408 | 2 | 6 | 22 | |
| GSE43470 | CRY2 | 1408 | 2 | 4 | 37 | |
| GSE46452 | CRY2 | 1408 | 8 | 5 | 46 | |
| GSE47630 | CRY2 | 1408 | 3 | 9 | 28 | |
| GSE54993 | CRY2 | 1408 | 3 | 0 | 67 | |
| GSE54994 | CRY2 | 1408 | 3 | 9 | 41 | |
| GSE60625 | CRY2 | 1408 | 0 | 0 | 11 | |
| GSE74703 | CRY2 | 1408 | 2 | 2 | 32 | |
| GSE74704 | CRY2 | 1408 | 1 | 4 | 15 | |
| TCGA | CRY2 | 1408 | 17 | 18 | 61 |
Total number of gains: 44; Total number of losses: 64; Total Number of normals: 380.
Somatic mutations of CRY2:
Generating mutation plots.
Highly correlated genes for CRY2:
Showing top 20/956 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRY2 | CHCHD10 | 0.781129 | 3 | 0 | 3 |
| CRY2 | SORCS1 | 0.751172 | 4 | 0 | 4 |
| CRY2 | CYP21A2 | 0.746579 | 3 | 0 | 3 |
| CRY2 | CLEC3B | 0.737106 | 3 | 0 | 3 |
| CRY2 | TMOD1 | 0.734596 | 5 | 0 | 5 |
| CRY2 | XKR4 | 0.714079 | 5 | 0 | 4 |
| CRY2 | PPP1R14A | 0.707939 | 4 | 0 | 4 |
| CRY2 | SGCG | 0.704611 | 5 | 0 | 5 |
| CRY2 | LSM11 | 0.697395 | 4 | 0 | 4 |
| CRY2 | CDX4 | 0.696188 | 4 | 0 | 4 |
| CRY2 | FBXO30 | 0.695302 | 4 | 0 | 3 |
| CRY2 | KCNB1 | 0.690079 | 5 | 0 | 5 |
| CRY2 | GPRASP1 | 0.689255 | 7 | 0 | 6 |
| CRY2 | GBA2 | 0.688242 | 5 | 0 | 3 |
| CRY2 | FAM104B | 0.684993 | 3 | 0 | 3 |
| CRY2 | FXYD1 | 0.683621 | 7 | 0 | 5 |
| CRY2 | PPP1R3B | 0.676949 | 3 | 0 | 3 |
| CRY2 | KCNN3 | 0.674582 | 5 | 0 | 4 |
| CRY2 | DDX42 | 0.673472 | 4 | 0 | 3 |
| CRY2 | PLIN4 | 0.672257 | 4 | 0 | 3 |
For details and further investigation, click here