| Full name: crystallin alpha B | Alias Symbol: HSPB5 | ||
| Type: protein-coding gene | Cytoband: 11q23.1 | ||
| Entrez ID: 1410 | HGNC ID: HGNC:2389 | Ensembl Gene: ENSG00000109846 | OMIM ID: 123590 |
| Drug and gene relationship at DGIdb | |||
CRYAB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04213 | Longevity regulating pathway - multiple species |
Expression of CRYAB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRYAB | 1410 | 209283_at | -1.4471 | 0.0462 | |
| GSE20347 | CRYAB | 1410 | 209283_at | -2.0286 | 0.0000 | |
| GSE23400 | CRYAB | 1410 | 209283_at | -1.7192 | 0.0000 | |
| GSE26886 | CRYAB | 1410 | 209283_at | -1.4385 | 0.0018 | |
| GSE29001 | CRYAB | 1410 | 209283_at | -2.2661 | 0.0002 | |
| GSE38129 | CRYAB | 1410 | 209283_at | -2.0023 | 0.0000 | |
| GSE45670 | CRYAB | 1410 | 209283_at | -0.8125 | 0.1744 | |
| GSE53622 | CRYAB | 1410 | 131032 | -2.0602 | 0.0000 | |
| GSE53624 | CRYAB | 1410 | 131032 | -2.0956 | 0.0000 | |
| GSE63941 | CRYAB | 1410 | 209283_at | -2.3151 | 0.0639 | |
| GSE77861 | CRYAB | 1410 | 209283_at | -1.0006 | 0.2710 | |
| GSE97050 | CRYAB | 1410 | A_24_P206776 | -1.6374 | 0.0700 | |
| SRP007169 | CRYAB | 1410 | RNAseq | -3.2153 | 0.0000 | |
| SRP008496 | CRYAB | 1410 | RNAseq | -2.9753 | 0.0002 | |
| SRP064894 | CRYAB | 1410 | RNAseq | -1.0280 | 0.0135 | |
| SRP133303 | CRYAB | 1410 | RNAseq | -1.5029 | 0.0043 | |
| SRP159526 | CRYAB | 1410 | RNAseq | -1.8743 | 0.0006 | |
| SRP193095 | CRYAB | 1410 | RNAseq | -1.6329 | 0.0000 | |
| SRP219564 | CRYAB | 1410 | RNAseq | -0.8651 | 0.3205 | |
| TCGA | CRYAB | 1410 | RNAseq | -0.3041 | 0.0636 |
Upregulated datasets: 0; Downregulated datasets: 14.
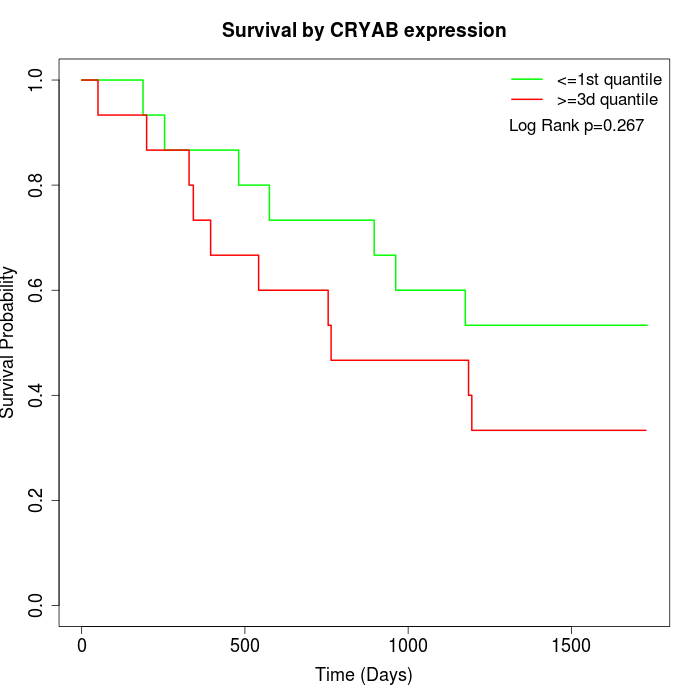
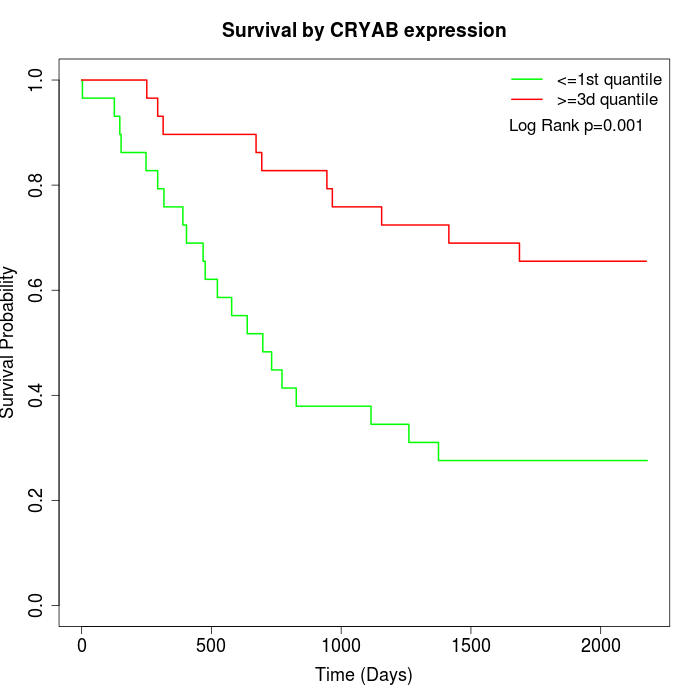
Survival by CRYAB expression:
Note: Click image to view full size file.
Copy number change of CRYAB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRYAB | 1410 | 0 | 12 | 18 | |
| GSE20123 | CRYAB | 1410 | 0 | 12 | 18 | |
| GSE43470 | CRYAB | 1410 | 2 | 6 | 35 | |
| GSE46452 | CRYAB | 1410 | 3 | 26 | 30 | |
| GSE47630 | CRYAB | 1410 | 2 | 19 | 19 | |
| GSE54993 | CRYAB | 1410 | 10 | 0 | 60 | |
| GSE54994 | CRYAB | 1410 | 5 | 19 | 29 | |
| GSE60625 | CRYAB | 1410 | 0 | 3 | 8 | |
| GSE74703 | CRYAB | 1410 | 1 | 4 | 31 | |
| GSE74704 | CRYAB | 1410 | 0 | 8 | 12 | |
| TCGA | CRYAB | 1410 | 7 | 48 | 41 |
Total number of gains: 30; Total number of losses: 157; Total Number of normals: 301.
Somatic mutations of CRYAB:
Generating mutation plots.
Highly correlated genes for CRYAB:
Showing top 20/1080 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRYAB | SORBS1 | 0.749684 | 4 | 0 | 4 |
| CRYAB | SPHKAP | 0.737105 | 4 | 0 | 4 |
| CRYAB | SLC25A25 | 0.734078 | 3 | 0 | 3 |
| CRYAB | PLCD4 | 0.724936 | 3 | 0 | 3 |
| CRYAB | CYP21A2 | 0.711228 | 3 | 0 | 3 |
| CRYAB | MYZAP | 0.70779 | 3 | 0 | 3 |
| CRYAB | HSPB8 | 0.70626 | 13 | 0 | 11 |
| CRYAB | C1QTNF7 | 0.698588 | 4 | 0 | 4 |
| CRYAB | RGS7BP | 0.696495 | 4 | 0 | 4 |
| CRYAB | RBFOX3 | 0.695742 | 3 | 0 | 3 |
| CRYAB | TIMP3 | 0.695564 | 3 | 0 | 3 |
| CRYAB | ASTN1 | 0.693013 | 3 | 0 | 3 |
| CRYAB | ARHGEF10L | 0.690667 | 11 | 0 | 11 |
| CRYAB | HMHB1 | 0.690329 | 3 | 0 | 3 |
| CRYAB | PINK1 | 0.684746 | 10 | 0 | 10 |
| CRYAB | MGLL | 0.682999 | 12 | 0 | 11 |
| CRYAB | ZNF365 | 0.678258 | 10 | 0 | 10 |
| CRYAB | PPP1R14A | 0.675975 | 4 | 0 | 4 |
| CRYAB | FOXP2 | 0.675097 | 4 | 0 | 4 |
| CRYAB | KRTAP5-10 | 0.673884 | 3 | 0 | 3 |
For details and further investigation, click here