| Full name: sorbin and SH3 domain containing 1 | Alias Symbol: FLJ12406|CAP|sh3p12|ponsin|KIAA1296 | ||
| Type: protein-coding gene | Cytoband: 10q24.1 | ||
| Entrez ID: 10580 | HGNC ID: HGNC:14565 | Ensembl Gene: ENSG00000095637 | OMIM ID: 605264 |
| Drug and gene relationship at DGIdb | |||
SORBS1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway | |
| hsa04520 | Adherens junction |
Expression of SORBS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SORBS1 | 10580 | 218087_s_at | -2.7117 | 0.1202 | |
| GSE20347 | SORBS1 | 10580 | 207436_x_at | 0.2053 | 0.2225 | |
| GSE23400 | SORBS1 | 10580 | 207436_x_at | -0.1909 | 0.0086 | |
| GSE26886 | SORBS1 | 10580 | 207436_x_at | 0.1095 | 0.3991 | |
| GSE29001 | SORBS1 | 10580 | 207436_x_at | -0.0188 | 0.9182 | |
| GSE38129 | SORBS1 | 10580 | 207436_x_at | -0.2456 | 0.1062 | |
| GSE45670 | SORBS1 | 10580 | 207436_x_at | -0.4051 | 0.0034 | |
| GSE53622 | SORBS1 | 10580 | 16689 | -2.6268 | 0.0000 | |
| GSE53624 | SORBS1 | 10580 | 44336 | -3.0952 | 0.0000 | |
| GSE63941 | SORBS1 | 10580 | 207436_x_at | 0.0476 | 0.7980 | |
| GSE77861 | SORBS1 | 10580 | 207436_x_at | 0.2388 | 0.3983 | |
| GSE97050 | SORBS1 | 10580 | A_33_P3288329 | -1.8045 | 0.0555 | |
| SRP007169 | SORBS1 | 10580 | RNAseq | -0.1425 | 0.7778 | |
| SRP008496 | SORBS1 | 10580 | RNAseq | 0.8536 | 0.1233 | |
| SRP064894 | SORBS1 | 10580 | RNAseq | -1.2244 | 0.0125 | |
| SRP133303 | SORBS1 | 10580 | RNAseq | -2.5074 | 0.0000 | |
| SRP159526 | SORBS1 | 10580 | RNAseq | -1.5249 | 0.0055 | |
| SRP193095 | SORBS1 | 10580 | RNAseq | -0.9516 | 0.0023 | |
| SRP219564 | SORBS1 | 10580 | RNAseq | -0.8878 | 0.3980 | |
| TCGA | SORBS1 | 10580 | RNAseq | -0.9471 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 5.
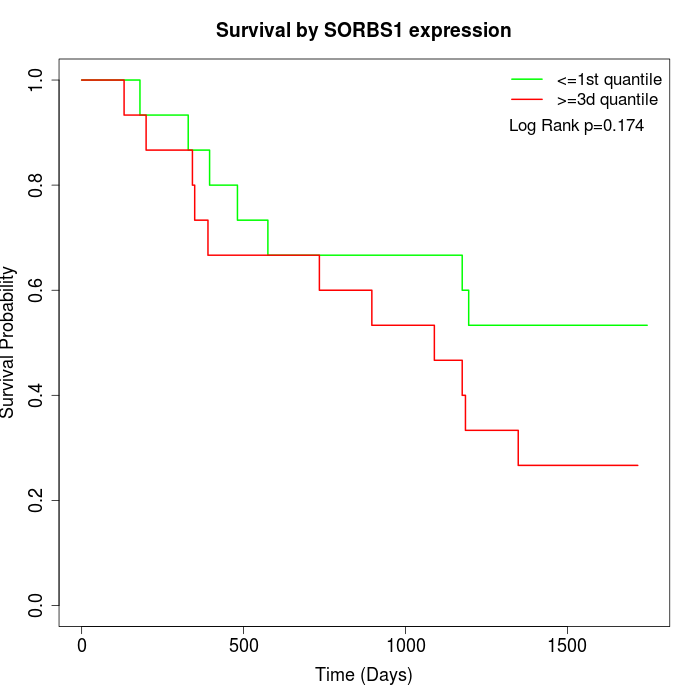
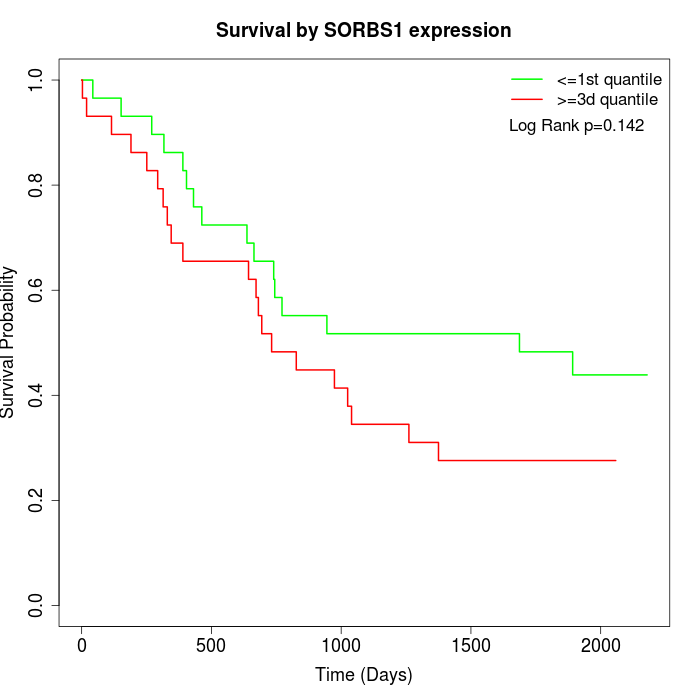
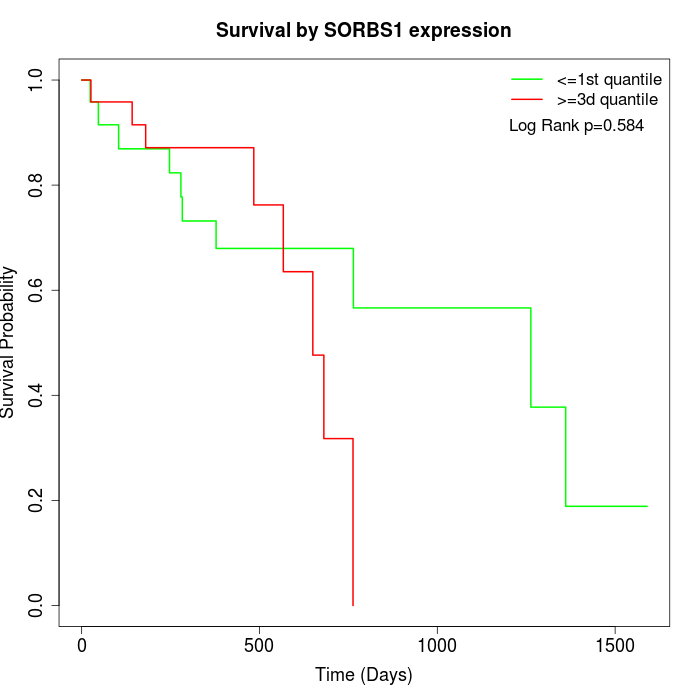
Survival by SORBS1 expression:
Note: Click image to view full size file.
Copy number change of SORBS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SORBS1 | 10580 | 1 | 8 | 21 | |
| GSE20123 | SORBS1 | 10580 | 1 | 7 | 22 | |
| GSE43470 | SORBS1 | 10580 | 0 | 7 | 36 | |
| GSE46452 | SORBS1 | 10580 | 0 | 11 | 48 | |
| GSE47630 | SORBS1 | 10580 | 2 | 14 | 24 | |
| GSE54993 | SORBS1 | 10580 | 7 | 0 | 63 | |
| GSE54994 | SORBS1 | 10580 | 2 | 10 | 41 | |
| GSE60625 | SORBS1 | 10580 | 0 | 0 | 11 | |
| GSE74703 | SORBS1 | 10580 | 0 | 5 | 31 | |
| GSE74704 | SORBS1 | 10580 | 0 | 4 | 16 | |
| TCGA | SORBS1 | 10580 | 6 | 27 | 63 |
Total number of gains: 19; Total number of losses: 93; Total Number of normals: 376.
Somatic mutations of SORBS1:
Generating mutation plots.
Highly correlated genes for SORBS1:
Showing top 20/676 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SORBS1 | RBPMS2 | 0.832747 | 6 | 0 | 6 |
| SORBS1 | LMOD1 | 0.831041 | 7 | 0 | 6 |
| SORBS1 | CYP21A2 | 0.815862 | 3 | 0 | 3 |
| SORBS1 | ASB5 | 0.811185 | 5 | 0 | 5 |
| SORBS1 | ASPA | 0.809508 | 5 | 0 | 5 |
| SORBS1 | ACTBL2 | 0.808719 | 3 | 0 | 3 |
| SORBS1 | UBXN10 | 0.804912 | 3 | 0 | 3 |
| SORBS1 | NEGR1 | 0.799873 | 5 | 0 | 5 |
| SORBS1 | PCP4 | 0.79946 | 7 | 0 | 6 |
| SORBS1 | LONRF2 | 0.796787 | 5 | 0 | 5 |
| SORBS1 | CLEC3B | 0.793662 | 3 | 0 | 3 |
| SORBS1 | RAB9B | 0.789915 | 6 | 0 | 6 |
| SORBS1 | ACTG2 | 0.788946 | 7 | 0 | 7 |
| SORBS1 | PDE5A | 0.786844 | 7 | 0 | 7 |
| SORBS1 | CASQ2 | 0.786773 | 7 | 0 | 7 |
| SORBS1 | SORCS1 | 0.77951 | 5 | 0 | 5 |
| SORBS1 | ANO5 | 0.778637 | 5 | 0 | 5 |
| SORBS1 | MRVI1 | 0.778314 | 6 | 0 | 5 |
| SORBS1 | TAOK1 | 0.777001 | 5 | 0 | 5 |
| SORBS1 | PLN | 0.77557 | 7 | 0 | 6 |
For details and further investigation, click here