| Full name: SPHK1 interactor, AKAP domain containing | Alias Symbol: SKIP | ||
| Type: protein-coding gene | Cytoband: 2q36.3 | ||
| Entrez ID: 80309 | HGNC ID: HGNC:30619 | Ensembl Gene: ENSG00000153820 | OMIM ID: 611646 |
| Drug and gene relationship at DGIdb | |||
Expression of SPHKAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPHKAP | 80309 | 228509_at | -0.3030 | 0.3004 | |
| GSE26886 | SPHKAP | 80309 | 228509_at | -0.1242 | 0.3606 | |
| GSE45670 | SPHKAP | 80309 | 228509_at | 0.0826 | 0.4064 | |
| GSE53622 | SPHKAP | 80309 | 28724 | -2.0488 | 0.0000 | |
| GSE53624 | SPHKAP | 80309 | 28724 | -2.2998 | 0.0000 | |
| GSE63941 | SPHKAP | 80309 | 228509_at | -0.0264 | 0.8976 | |
| GSE77861 | SPHKAP | 80309 | 228509_at | -0.2169 | 0.1185 | |
| TCGA | SPHKAP | 80309 | RNAseq | -3.1335 | 0.0049 |
Upregulated datasets: 0; Downregulated datasets: 3.
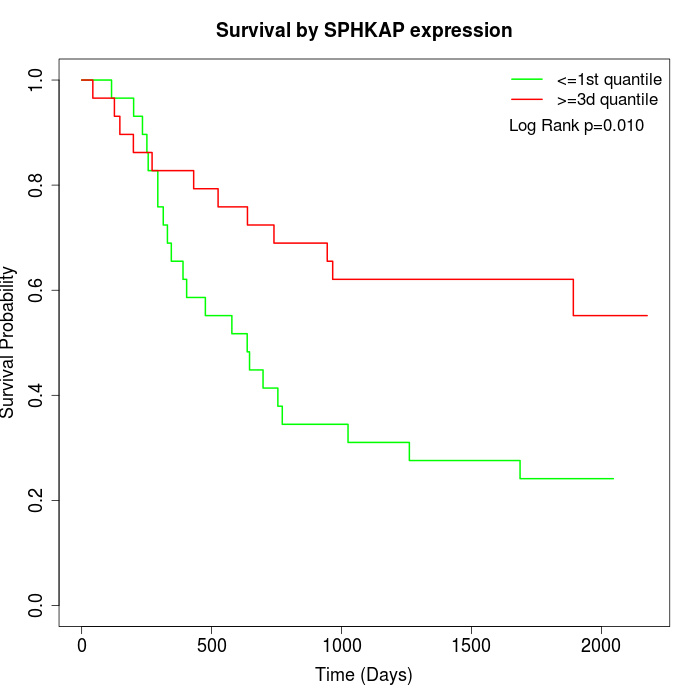
Survival by SPHKAP expression:
Note: Click image to view full size file.
Copy number change of SPHKAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPHKAP | 80309 | 2 | 12 | 16 | |
| GSE20123 | SPHKAP | 80309 | 2 | 12 | 16 | |
| GSE43470 | SPHKAP | 80309 | 2 | 7 | 34 | |
| GSE46452 | SPHKAP | 80309 | 0 | 5 | 54 | |
| GSE47630 | SPHKAP | 80309 | 4 | 5 | 31 | |
| GSE54993 | SPHKAP | 80309 | 1 | 2 | 67 | |
| GSE54994 | SPHKAP | 80309 | 6 | 11 | 36 | |
| GSE60625 | SPHKAP | 80309 | 0 | 3 | 8 | |
| GSE74703 | SPHKAP | 80309 | 2 | 6 | 28 | |
| GSE74704 | SPHKAP | 80309 | 2 | 6 | 12 | |
| TCGA | SPHKAP | 80309 | 13 | 26 | 57 |
Total number of gains: 34; Total number of losses: 95; Total Number of normals: 359.
Somatic mutations of SPHKAP:
Generating mutation plots.
Highly correlated genes for SPHKAP:
Showing top 20/538 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPHKAP | ANXA1 | 0.779711 | 3 | 0 | 3 |
| SPHKAP | MALL | 0.777774 | 3 | 0 | 3 |
| SPHKAP | TMOD3 | 0.777105 | 3 | 0 | 3 |
| SPHKAP | CNFN | 0.76693 | 3 | 0 | 3 |
| SPHKAP | SERPINB1 | 0.765141 | 3 | 0 | 3 |
| SPHKAP | SFRP5 | 0.759396 | 4 | 0 | 4 |
| SPHKAP | EPS8L1 | 0.754164 | 3 | 0 | 3 |
| SPHKAP | PPL | 0.754091 | 3 | 0 | 3 |
| SPHKAP | SCEL | 0.752756 | 3 | 0 | 3 |
| SPHKAP | SASH1 | 0.747331 | 3 | 0 | 3 |
| SPHKAP | RANBP9 | 0.745748 | 3 | 0 | 3 |
| SPHKAP | MASP2 | 0.742606 | 3 | 0 | 3 |
| SPHKAP | PHACTR2 | 0.741289 | 3 | 0 | 3 |
| SPHKAP | MXD1 | 0.740587 | 3 | 0 | 3 |
| SPHKAP | C18orf25 | 0.738148 | 3 | 0 | 3 |
| SPHKAP | CRYAB | 0.737105 | 4 | 0 | 4 |
| SPHKAP | MPZL3 | 0.736554 | 3 | 0 | 3 |
| SPHKAP | TMPRSS11E | 0.734397 | 3 | 0 | 3 |
| SPHKAP | C5orf66-AS1 | 0.729366 | 3 | 0 | 3 |
| SPHKAP | ABLIM1 | 0.72904 | 3 | 0 | 3 |
For details and further investigation, click here