| Full name: casein kinase 1 gamma 2 | Alias Symbol: CK1g2 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 1455 | HGNC ID: HGNC:2455 | Ensembl Gene: ENSG00000133275 | OMIM ID: 602214 |
| Drug and gene relationship at DGIdb | |||
CSNK1G2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04340 | Hedgehog signaling pathway |
Expression of CSNK1G2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSNK1G2 | 1455 | 202573_at | 0.2322 | 0.4025 | |
| GSE20347 | CSNK1G2 | 1455 | 202573_at | 0.3076 | 0.0202 | |
| GSE23400 | CSNK1G2 | 1455 | 202573_at | 0.1726 | 0.0023 | |
| GSE26886 | CSNK1G2 | 1455 | 202573_at | 0.9813 | 0.0000 | |
| GSE29001 | CSNK1G2 | 1455 | 202573_at | 0.2145 | 0.5462 | |
| GSE38129 | CSNK1G2 | 1455 | 202573_at | 0.3626 | 0.0007 | |
| GSE45670 | CSNK1G2 | 1455 | 202573_at | 0.2842 | 0.0043 | |
| GSE53622 | CSNK1G2 | 1455 | 81715 | 0.5518 | 0.0000 | |
| GSE53624 | CSNK1G2 | 1455 | 81715 | 0.5936 | 0.0000 | |
| GSE63941 | CSNK1G2 | 1455 | 202573_at | -0.4537 | 0.0948 | |
| GSE77861 | CSNK1G2 | 1455 | 202573_at | 0.4054 | 0.0149 | |
| GSE97050 | CSNK1G2 | 1455 | A_33_P3259017 | 0.5888 | 0.1404 | |
| SRP007169 | CSNK1G2 | 1455 | RNAseq | -0.2899 | 0.3569 | |
| SRP008496 | CSNK1G2 | 1455 | RNAseq | -0.3944 | 0.1043 | |
| SRP064894 | CSNK1G2 | 1455 | RNAseq | 0.3423 | 0.0582 | |
| SRP133303 | CSNK1G2 | 1455 | RNAseq | 0.4213 | 0.0235 | |
| SRP159526 | CSNK1G2 | 1455 | RNAseq | 0.2546 | 0.3178 | |
| SRP193095 | CSNK1G2 | 1455 | RNAseq | 0.4244 | 0.0005 | |
| SRP219564 | CSNK1G2 | 1455 | RNAseq | 0.1110 | 0.6652 | |
| TCGA | CSNK1G2 | 1455 | RNAseq | 0.1472 | 0.0017 |
Upregulated datasets: 0; Downregulated datasets: 0.
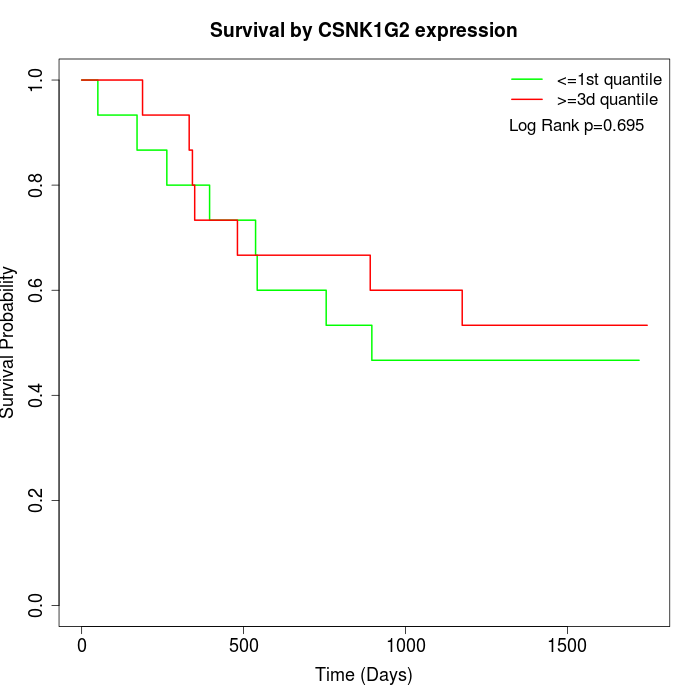
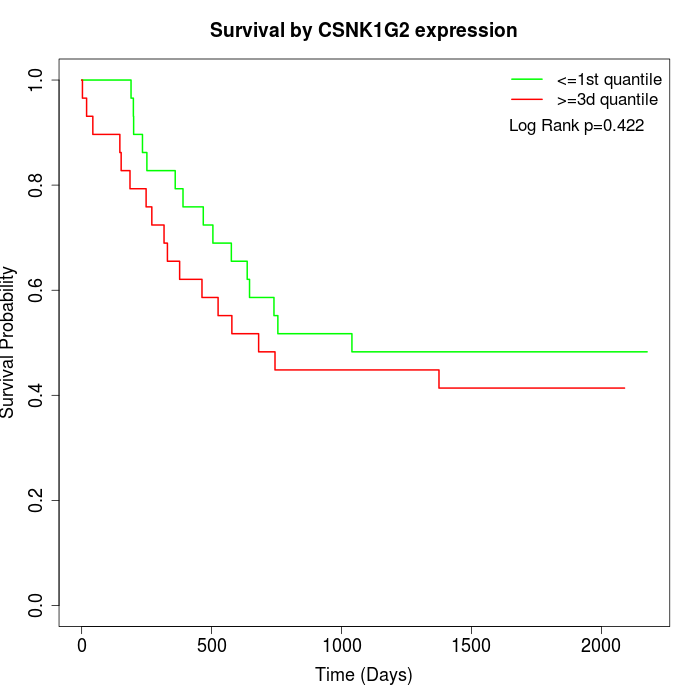
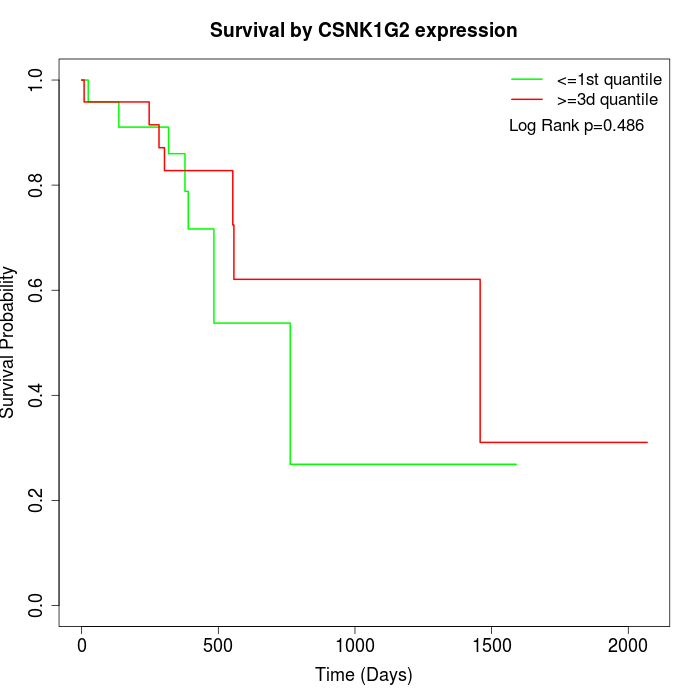
Survival by CSNK1G2 expression:
Note: Click image to view full size file.
Copy number change of CSNK1G2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSNK1G2 | 1455 | 4 | 6 | 20 | |
| GSE20123 | CSNK1G2 | 1455 | 4 | 5 | 21 | |
| GSE43470 | CSNK1G2 | 1455 | 1 | 10 | 32 | |
| GSE46452 | CSNK1G2 | 1455 | 47 | 1 | 11 | |
| GSE47630 | CSNK1G2 | 1455 | 5 | 7 | 28 | |
| GSE54993 | CSNK1G2 | 1455 | 16 | 3 | 51 | |
| GSE54994 | CSNK1G2 | 1455 | 8 | 15 | 30 | |
| GSE60625 | CSNK1G2 | 1455 | 9 | 0 | 2 | |
| GSE74703 | CSNK1G2 | 1455 | 1 | 7 | 28 | |
| GSE74704 | CSNK1G2 | 1455 | 3 | 3 | 14 | |
| TCGA | CSNK1G2 | 1455 | 6 | 25 | 65 |
Total number of gains: 104; Total number of losses: 82; Total Number of normals: 302.
Somatic mutations of CSNK1G2:
Generating mutation plots.
Highly correlated genes for CSNK1G2:
Showing top 20/1019 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSNK1G2 | C1orf122 | 0.799986 | 3 | 0 | 3 |
| CSNK1G2 | DAPK3 | 0.796641 | 3 | 0 | 3 |
| CSNK1G2 | FCHO1 | 0.783877 | 4 | 0 | 4 |
| CSNK1G2 | DGCR2 | 0.775571 | 4 | 0 | 4 |
| CSNK1G2 | B3GALT6 | 0.753083 | 6 | 0 | 6 |
| CSNK1G2 | DBN1 | 0.751643 | 4 | 0 | 4 |
| CSNK1G2 | C8orf76 | 0.749202 | 4 | 0 | 4 |
| CSNK1G2 | SARDH | 0.746895 | 3 | 0 | 3 |
| CSNK1G2 | DNAJB11 | 0.745743 | 5 | 0 | 5 |
| CSNK1G2 | TMEM189 | 0.744059 | 3 | 0 | 3 |
| CSNK1G2 | RIOK1 | 0.73697 | 5 | 0 | 5 |
| CSNK1G2 | ACAD9 | 0.735972 | 3 | 0 | 3 |
| CSNK1G2 | XXYLT1 | 0.73445 | 5 | 0 | 4 |
| CSNK1G2 | COPB1 | 0.728983 | 5 | 0 | 4 |
| CSNK1G2 | SRA1 | 0.725936 | 3 | 0 | 3 |
| CSNK1G2 | WDYHV1 | 0.723575 | 4 | 0 | 4 |
| CSNK1G2 | OPLAH | 0.723107 | 3 | 0 | 3 |
| CSNK1G2 | PMS2 | 0.721741 | 3 | 0 | 3 |
| CSNK1G2 | PYCR2 | 0.721196 | 3 | 0 | 3 |
| CSNK1G2 | GATAD2A | 0.721064 | 6 | 0 | 6 |
For details and further investigation, click here