| Full name: cystatin SA | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 20p11.21 | ||
| Entrez ID: 1470 | HGNC ID: HGNC:2474 | Ensembl Gene: ENSG00000170369 | OMIM ID: 123856 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CST2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CST2 | 1470 | 208555_x_at | 1.0112 | 0.0726 | |
| GSE20347 | CST2 | 1470 | 208555_x_at | 0.3442 | 0.0042 | |
| GSE23400 | CST2 | 1470 | 208555_x_at | 0.3817 | 0.0000 | |
| GSE26886 | CST2 | 1470 | 208555_x_at | 0.4465 | 0.0027 | |
| GSE29001 | CST2 | 1470 | 208555_x_at | -0.0550 | 0.7380 | |
| GSE38129 | CST2 | 1470 | 208555_x_at | 0.9355 | 0.0000 | |
| GSE45670 | CST2 | 1470 | 208555_x_at | 0.1201 | 0.3836 | |
| GSE53622 | CST2 | 1470 | 13386 | 5.0836 | 0.0000 | |
| GSE53624 | CST2 | 1470 | 13386 | 4.3895 | 0.0000 | |
| GSE63941 | CST2 | 1470 | 208555_x_at | 0.2765 | 0.1302 | |
| GSE77861 | CST2 | 1470 | 208555_x_at | -0.1539 | 0.3222 | |
| GSE97050 | CST2 | 1470 | A_24_P237175 | 2.8068 | 0.1058 | |
| SRP219564 | CST2 | 1470 | RNAseq | 3.8367 | 0.0040 | |
| TCGA | CST2 | 1470 | RNAseq | 1.8325 | 0.0009 |
Upregulated datasets: 4; Downregulated datasets: 0.
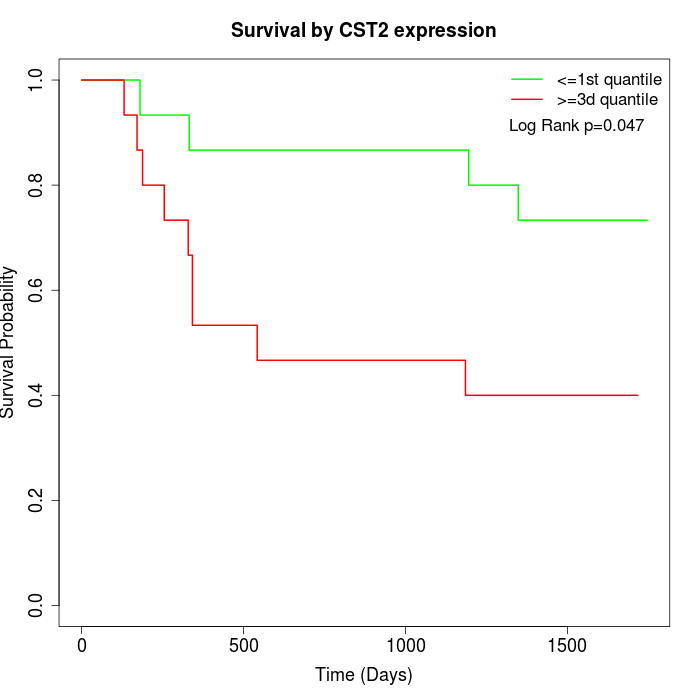
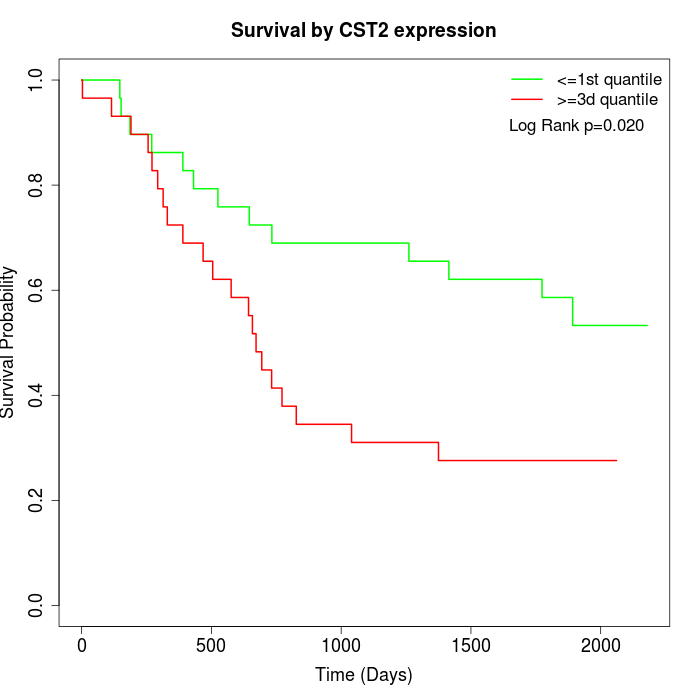
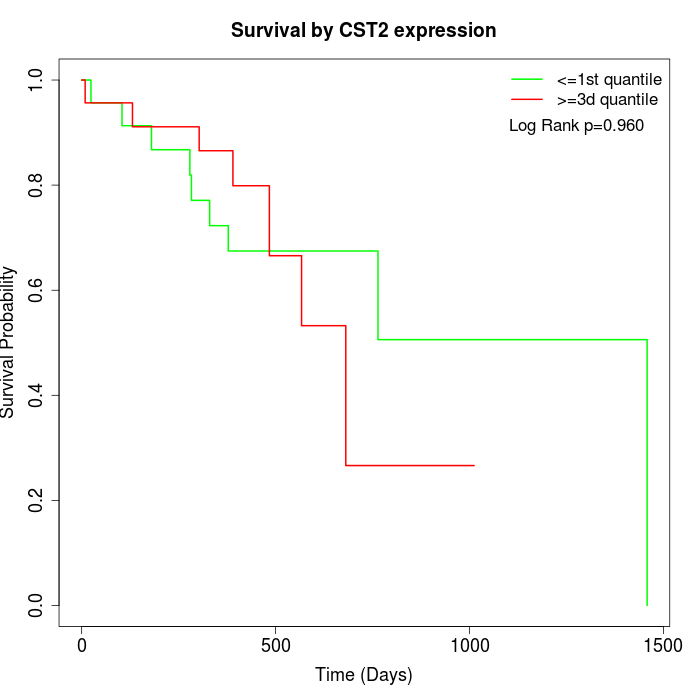
Survival by CST2 expression:
Note: Click image to view full size file.
Copy number change of CST2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CST2 | 1470 | 12 | 2 | 16 | |
| GSE20123 | CST2 | 1470 | 12 | 2 | 16 | |
| GSE43470 | CST2 | 1470 | 12 | 1 | 30 | |
| GSE46452 | CST2 | 1470 | 27 | 1 | 31 | |
| GSE47630 | CST2 | 1470 | 17 | 4 | 19 | |
| GSE54993 | CST2 | 1470 | 0 | 17 | 53 | |
| GSE54994 | CST2 | 1470 | 27 | 1 | 25 | |
| GSE60625 | CST2 | 1470 | 0 | 0 | 11 | |
| GSE74703 | CST2 | 1470 | 10 | 1 | 25 | |
| GSE74704 | CST2 | 1470 | 7 | 1 | 12 | |
| TCGA | CST2 | 1470 | 43 | 10 | 43 |
Total number of gains: 167; Total number of losses: 40; Total Number of normals: 281.
Somatic mutations of CST2:
Generating mutation plots.
Highly correlated genes for CST2:
Showing top 20/594 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CST2 | CST1 | 0.757115 | 11 | 0 | 9 |
| CST2 | CST4 | 0.725394 | 8 | 0 | 7 |
| CST2 | AMN | 0.719406 | 3 | 0 | 3 |
| CST2 | PNPLA6 | 0.705406 | 3 | 0 | 3 |
| CST2 | COL10A1 | 0.69939 | 9 | 0 | 8 |
| CST2 | TANGO2 | 0.699037 | 3 | 0 | 3 |
| CST2 | SNAP47 | 0.696253 | 3 | 0 | 3 |
| CST2 | BRAT1 | 0.69506 | 3 | 0 | 3 |
| CST2 | YDJC | 0.686278 | 5 | 0 | 5 |
| CST2 | TAB1 | 0.686036 | 3 | 0 | 3 |
| CST2 | KREMEN2 | 0.682796 | 3 | 0 | 3 |
| CST2 | SLC5A12 | 0.671307 | 6 | 0 | 6 |
| CST2 | HAGLROS | 0.665153 | 4 | 0 | 4 |
| CST2 | CPAMD8 | 0.664934 | 3 | 0 | 3 |
| CST2 | COL9A2 | 0.6631 | 5 | 0 | 4 |
| CST2 | PLXNA1 | 0.662442 | 7 | 0 | 7 |
| CST2 | NFKBIL1 | 0.66168 | 3 | 0 | 3 |
| CST2 | FANCB | 0.661211 | 3 | 0 | 3 |
| CST2 | SLC39A3 | 0.660537 | 4 | 0 | 3 |
| CST2 | SIX1 | 0.658182 | 8 | 0 | 6 |
For details and further investigation, click here