| Full name: cystatin S | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 20p11.21 | ||
| Entrez ID: 1472 | HGNC ID: HGNC:2476 | Ensembl Gene: ENSG00000101441 | OMIM ID: 123857 |
| Drug and gene relationship at DGIdb | |||
Expression of CST4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CST4 | 1472 | 206994_at | 1.5275 | 0.0843 | |
| GSE20347 | CST4 | 1472 | 206994_at | 0.5765 | 0.0055 | |
| GSE23400 | CST4 | 1472 | 206994_at | 0.6152 | 0.0000 | |
| GSE26886 | CST4 | 1472 | 206994_at | 0.3582 | 0.0924 | |
| GSE29001 | CST4 | 1472 | 206994_at | 0.3468 | 0.0612 | |
| GSE38129 | CST4 | 1472 | 206994_at | 1.2821 | 0.0000 | |
| GSE45670 | CST4 | 1472 | 206994_at | 0.2536 | 0.0606 | |
| GSE63941 | CST4 | 1472 | 206994_at | -0.5223 | 0.0052 | |
| GSE77861 | CST4 | 1472 | 206994_at | -0.1133 | 0.5071 | |
| TCGA | CST4 | 1472 | RNAseq | 5.1347 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
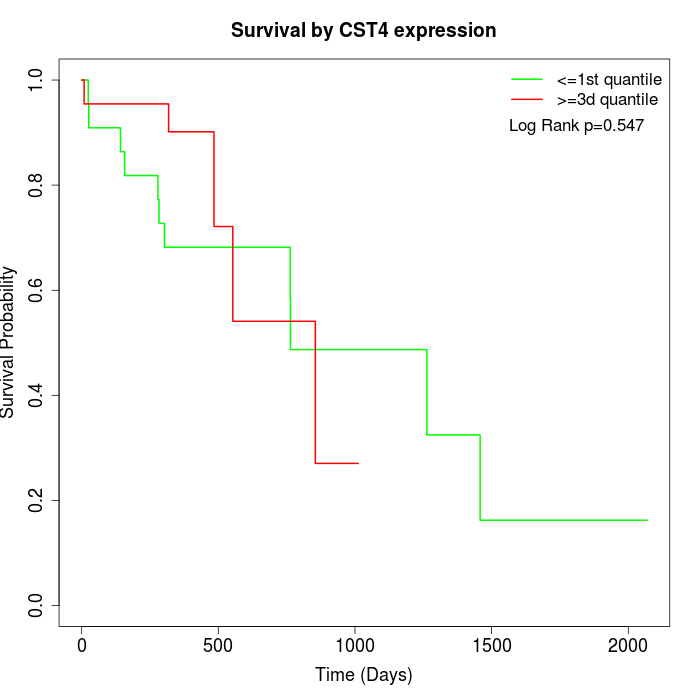
Survival by CST4 expression:
Note: Click image to view full size file.
Copy number change of CST4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CST4 | 1472 | 12 | 2 | 16 | |
| GSE20123 | CST4 | 1472 | 12 | 2 | 16 | |
| GSE43470 | CST4 | 1472 | 12 | 1 | 30 | |
| GSE46452 | CST4 | 1472 | 27 | 1 | 31 | |
| GSE47630 | CST4 | 1472 | 17 | 4 | 19 | |
| GSE54993 | CST4 | 1472 | 0 | 17 | 53 | |
| GSE54994 | CST4 | 1472 | 26 | 2 | 25 | |
| GSE60625 | CST4 | 1472 | 0 | 0 | 11 | |
| GSE74703 | CST4 | 1472 | 10 | 1 | 25 | |
| GSE74704 | CST4 | 1472 | 7 | 1 | 12 | |
| TCGA | CST4 | 1472 | 43 | 10 | 43 |
Total number of gains: 166; Total number of losses: 41; Total Number of normals: 281.
Somatic mutations of CST4:
Generating mutation plots.
Highly correlated genes for CST4:
Showing top 20/154 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CST4 | CST1 | 0.823432 | 8 | 0 | 8 |
| CST4 | CST2 | 0.725394 | 8 | 0 | 7 |
| CST4 | FOXP4 | 0.725352 | 3 | 0 | 3 |
| CST4 | CBS | 0.670801 | 4 | 0 | 4 |
| CST4 | THAP7 | 0.66393 | 3 | 0 | 3 |
| CST4 | MMP11 | 0.659777 | 7 | 0 | 6 |
| CST4 | ZNF74 | 0.659689 | 4 | 0 | 3 |
| CST4 | WDR83 | 0.659446 | 3 | 0 | 3 |
| CST4 | LRRC45 | 0.656051 | 3 | 0 | 3 |
| CST4 | BAHD1 | 0.651938 | 4 | 0 | 4 |
| CST4 | ATOH8 | 0.651208 | 3 | 0 | 3 |
| CST4 | DCUN1D2 | 0.643744 | 3 | 0 | 3 |
| CST4 | CD70 | 0.64323 | 3 | 0 | 3 |
| CST4 | TEAD2 | 0.635268 | 3 | 0 | 3 |
| CST4 | MAP1S | 0.634964 | 5 | 0 | 3 |
| CST4 | MARCKSL1 | 0.634071 | 5 | 0 | 4 |
| CST4 | COL10A1 | 0.630582 | 7 | 0 | 6 |
| CST4 | RPGRIP1L | 0.62656 | 4 | 0 | 3 |
| CST4 | SUSD5 | 0.610345 | 4 | 0 | 4 |
| CST4 | EVA1B | 0.607029 | 4 | 0 | 3 |
For details and further investigation, click here