| Full name: cut like homeobox 2 | Alias Symbol: KIAA0293|CDP2 | ||
| Type: protein-coding gene | Cytoband: 12q24.11-q24.12 | ||
| Entrez ID: 23316 | HGNC ID: HGNC:19347 | Ensembl Gene: ENSG00000111249 | OMIM ID: 610648 |
| Drug and gene relationship at DGIdb | |||
Expression of CUX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CUX2 | 23316 | 1566528_at | 0.1126 | 0.5777 | |
| GSE20347 | CUX2 | 23316 | 213920_at | 0.0019 | 0.9766 | |
| GSE23400 | CUX2 | 23316 | 213920_at | -0.0462 | 0.0193 | |
| GSE26886 | CUX2 | 23316 | 1566528_at | -0.0534 | 0.6977 | |
| GSE29001 | CUX2 | 23316 | 213920_at | 0.1967 | 0.5431 | |
| GSE38129 | CUX2 | 23316 | 213920_at | 0.1389 | 0.6305 | |
| GSE45670 | CUX2 | 23316 | 1566528_at | 0.0138 | 0.9012 | |
| GSE53622 | CUX2 | 23316 | 72977 | -1.3875 | 0.0000 | |
| GSE53624 | CUX2 | 23316 | 84673 | 0.2252 | 0.3127 | |
| GSE63941 | CUX2 | 23316 | 1566528_at | -0.0247 | 0.8490 | |
| GSE77861 | CUX2 | 23316 | 1566528_at | -0.1903 | 0.1848 | |
| TCGA | CUX2 | 23316 | RNAseq | -2.8038 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 2.
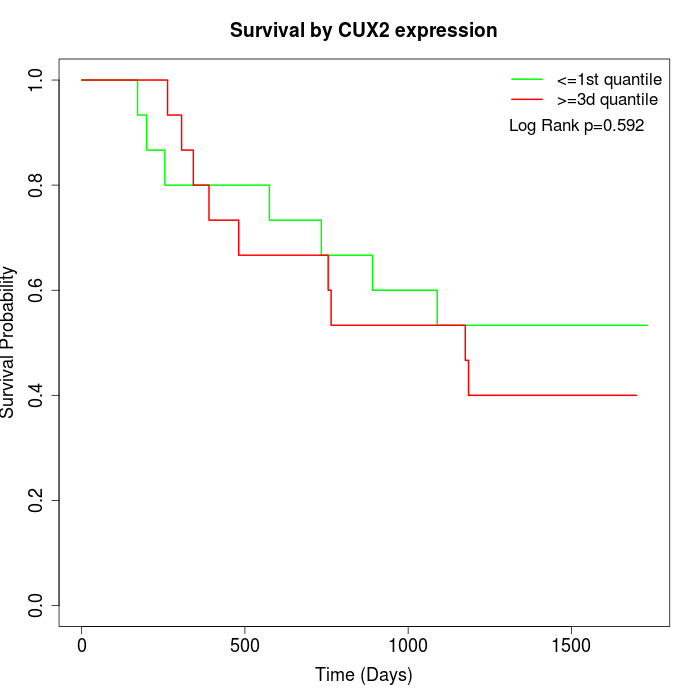
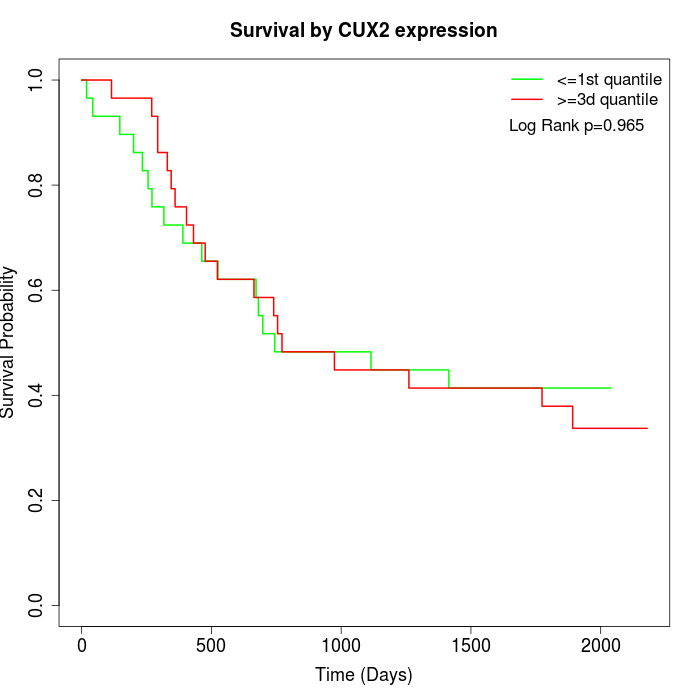
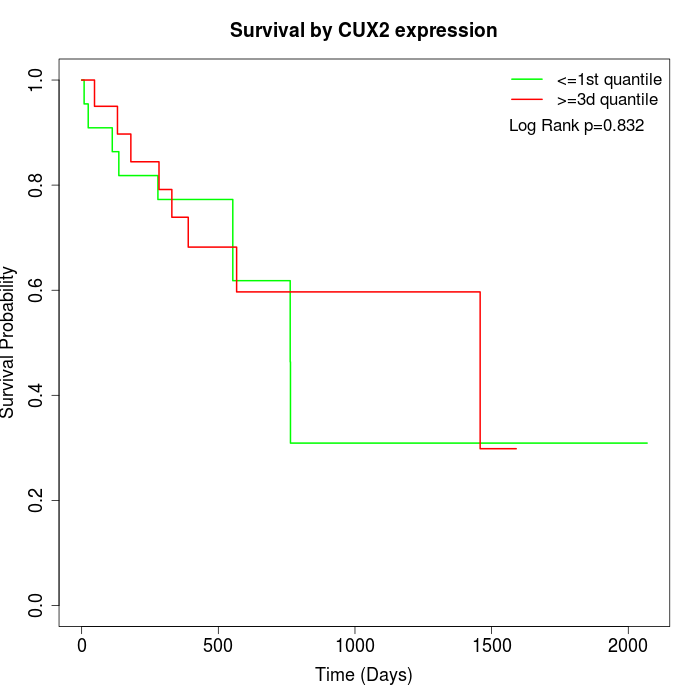
Survival by CUX2 expression:
Note: Click image to view full size file.
Copy number change of CUX2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CUX2 | 23316 | 5 | 3 | 22 | |
| GSE20123 | CUX2 | 23316 | 5 | 3 | 22 | |
| GSE43470 | CUX2 | 23316 | 2 | 1 | 40 | |
| GSE46452 | CUX2 | 23316 | 9 | 1 | 49 | |
| GSE47630 | CUX2 | 23316 | 9 | 3 | 28 | |
| GSE54993 | CUX2 | 23316 | 0 | 5 | 65 | |
| GSE54994 | CUX2 | 23316 | 5 | 2 | 46 | |
| GSE60625 | CUX2 | 23316 | 0 | 0 | 11 | |
| GSE74703 | CUX2 | 23316 | 2 | 0 | 34 | |
| GSE74704 | CUX2 | 23316 | 2 | 2 | 16 | |
| TCGA | CUX2 | 23316 | 22 | 11 | 63 |
Total number of gains: 61; Total number of losses: 31; Total Number of normals: 396.
Somatic mutations of CUX2:
Generating mutation plots.
Highly correlated genes for CUX2:
Showing top 20/58 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CUX2 | APOBEC2 | 0.678979 | 3 | 0 | 3 |
| CUX2 | CGA | 0.672879 | 3 | 0 | 3 |
| CUX2 | MAGEC2 | 0.644668 | 4 | 0 | 3 |
| CUX2 | NCR1 | 0.640651 | 5 | 0 | 4 |
| CUX2 | FGF5 | 0.628196 | 4 | 0 | 3 |
| CUX2 | WFIKKN2 | 0.625784 | 3 | 0 | 3 |
| CUX2 | HCCAT5 | 0.618096 | 3 | 0 | 3 |
| CUX2 | ZNF547 | 0.601636 | 3 | 0 | 3 |
| CUX2 | TMPRSS5 | 0.601425 | 3 | 0 | 3 |
| CUX2 | CALCA | 0.595557 | 5 | 0 | 4 |
| CUX2 | DCD | 0.593644 | 4 | 0 | 3 |
| CUX2 | APOA1 | 0.58507 | 4 | 0 | 3 |
| CUX2 | SUN5 | 0.584537 | 4 | 0 | 4 |
| CUX2 | P2RX3 | 0.583295 | 4 | 0 | 3 |
| CUX2 | SCTR | 0.58156 | 3 | 0 | 3 |
| CUX2 | ADAMTS7 | 0.569281 | 4 | 0 | 3 |
| CUX2 | ATP2B2 | 0.567617 | 4 | 0 | 3 |
| CUX2 | BRINP2 | 0.56703 | 4 | 0 | 3 |
| CUX2 | UPB1 | 0.563816 | 6 | 0 | 4 |
| CUX2 | CNTNAP5 | 0.562654 | 3 | 0 | 3 |
For details and further investigation, click here