| Full name: cytochrome P450 family 2 subfamily A member 13 | Alias Symbol: CPAD|CYP2A | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 1553 | HGNC ID: HGNC:2608 | Ensembl Gene: ENSG00000197838 | OMIM ID: 608055 |
| Drug and gene relationship at DGIdb | |||
Expression of CYP2A13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CYP2A13 | 1553 | 208327_at | 0.0896 | 0.7134 | |
| GSE20347 | CYP2A13 | 1553 | 208327_at | 0.0087 | 0.9289 | |
| GSE23400 | CYP2A13 | 1553 | 208327_at | -0.0514 | 0.1673 | |
| GSE26886 | CYP2A13 | 1553 | 208327_at | 0.2656 | 0.0293 | |
| GSE29001 | CYP2A13 | 1553 | 208327_at | -0.2103 | 0.2126 | |
| GSE38129 | CYP2A13 | 1553 | 208327_at | -0.0877 | 0.2386 | |
| GSE45670 | CYP2A13 | 1553 | 208327_at | -0.1681 | 0.0935 | |
| GSE53622 | CYP2A13 | 1553 | 97656 | 0.3864 | 0.0012 | |
| GSE53624 | CYP2A13 | 1553 | 97656 | 0.3563 | 0.0003 | |
| GSE63941 | CYP2A13 | 1553 | 208327_at | 0.4875 | 0.0087 | |
| GSE77861 | CYP2A13 | 1553 | 208327_at | 0.0018 | 0.9943 |
Upregulated datasets: 0; Downregulated datasets: 0.
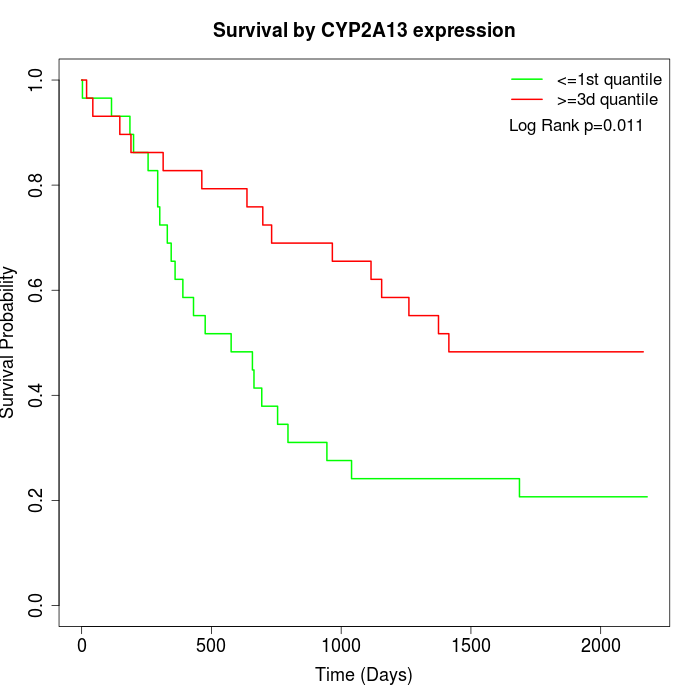
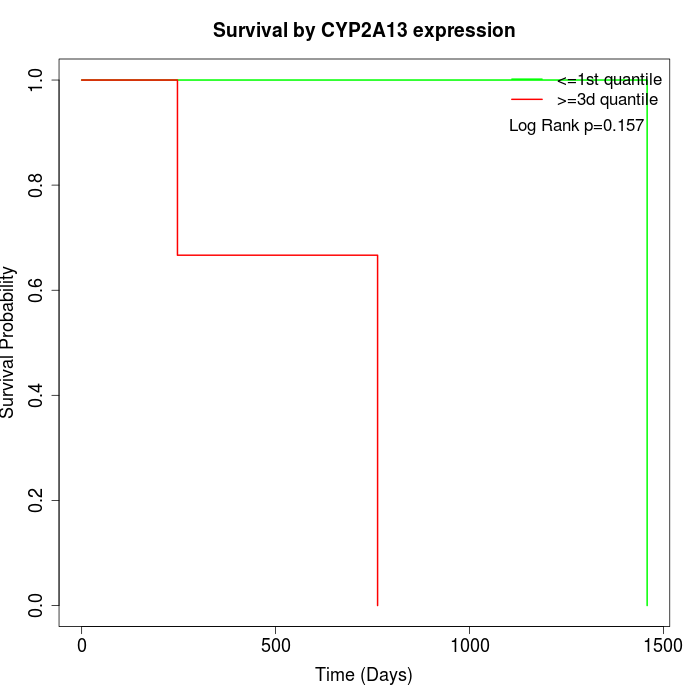
Survival by CYP2A13 expression:
Note: Click image to view full size file.
Copy number change of CYP2A13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CYP2A13 | 1553 | 4 | 4 | 22 | |
| GSE20123 | CYP2A13 | 1553 | 4 | 3 | 23 | |
| GSE43470 | CYP2A13 | 1553 | 3 | 11 | 29 | |
| GSE46452 | CYP2A13 | 1553 | 48 | 1 | 10 | |
| GSE47630 | CYP2A13 | 1553 | 9 | 5 | 26 | |
| GSE54993 | CYP2A13 | 1553 | 17 | 4 | 49 | |
| GSE54994 | CYP2A13 | 1553 | 6 | 11 | 36 | |
| GSE60625 | CYP2A13 | 1553 | 9 | 0 | 2 | |
| GSE74703 | CYP2A13 | 1553 | 3 | 7 | 26 | |
| GSE74704 | CYP2A13 | 1553 | 4 | 1 | 15 | |
| TCGA | CYP2A13 | 1553 | 14 | 16 | 66 |
Total number of gains: 121; Total number of losses: 63; Total Number of normals: 304.
Somatic mutations of CYP2A13:
Generating mutation plots.
Highly correlated genes for CYP2A13:
Showing top 20/482 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CYP2A13 | DGCR9 | 0.717698 | 3 | 0 | 3 |
| CYP2A13 | KCNK16 | 0.710178 | 3 | 0 | 3 |
| CYP2A13 | TMEM63A | 0.693183 | 4 | 0 | 4 |
| CYP2A13 | SRL | 0.682043 | 3 | 0 | 3 |
| CYP2A13 | ZNF428 | 0.681152 | 3 | 0 | 3 |
| CYP2A13 | CNIH2 | 0.67741 | 3 | 0 | 3 |
| CYP2A13 | KDM5C | 0.674629 | 4 | 0 | 4 |
| CYP2A13 | TRIM51 | 0.674305 | 3 | 0 | 3 |
| CYP2A13 | NKX2-2 | 0.667279 | 3 | 0 | 3 |
| CYP2A13 | R3HDM4 | 0.665775 | 3 | 0 | 3 |
| CYP2A13 | CNTD2 | 0.66495 | 5 | 0 | 5 |
| CYP2A13 | MYO15B | 0.661698 | 3 | 0 | 3 |
| CYP2A13 | NAT2 | 0.654107 | 3 | 0 | 3 |
| CYP2A13 | RPL38 | 0.646862 | 3 | 0 | 3 |
| CYP2A13 | TRAF2 | 0.646821 | 3 | 0 | 3 |
| CYP2A13 | ZNF771 | 0.645838 | 6 | 0 | 5 |
| CYP2A13 | PRSS53 | 0.642878 | 3 | 0 | 3 |
| CYP2A13 | TMEM102 | 0.637723 | 4 | 0 | 3 |
| CYP2A13 | RAB3A | 0.636354 | 5 | 0 | 4 |
| CYP2A13 | RAB4B | 0.63585 | 3 | 0 | 3 |
For details and further investigation, click here