| Full name: potassium two pore domain channel subfamily K member 16 | Alias Symbol: K2p16.1|TALK-1|TALK1 | ||
| Type: protein-coding gene | Cytoband: 6p21.2 | ||
| Entrez ID: 83795 | HGNC ID: HGNC:14464 | Ensembl Gene: ENSG00000095981 | OMIM ID: 607369 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNK16:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNK16 | 83795 | 234554_at | 0.0022 | 0.9955 | |
| GSE26886 | KCNK16 | 83795 | 234554_at | 0.1736 | 0.1239 | |
| GSE45670 | KCNK16 | 83795 | 234554_at | 0.0681 | 0.4741 | |
| GSE53622 | KCNK16 | 83795 | 70775 | 0.0718 | 0.6722 | |
| GSE53624 | KCNK16 | 83795 | 70775 | -0.0813 | 0.3147 | |
| GSE63941 | KCNK16 | 83795 | 234554_at | 0.2197 | 0.1863 | |
| GSE77861 | KCNK16 | 83795 | 234554_at | -0.0061 | 0.9764 | |
| TCGA | KCNK16 | 83795 | RNAseq | -4.0176 | 0.1005 |
Upregulated datasets: 0; Downregulated datasets: 0.
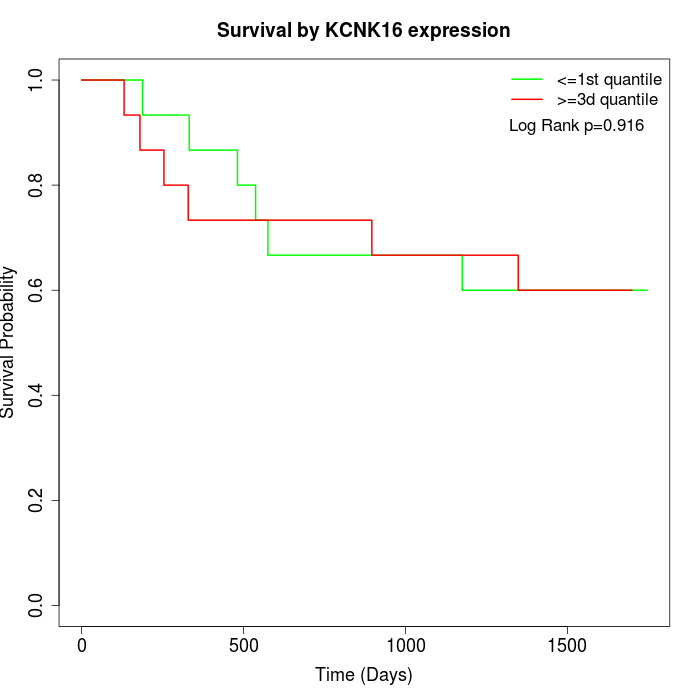
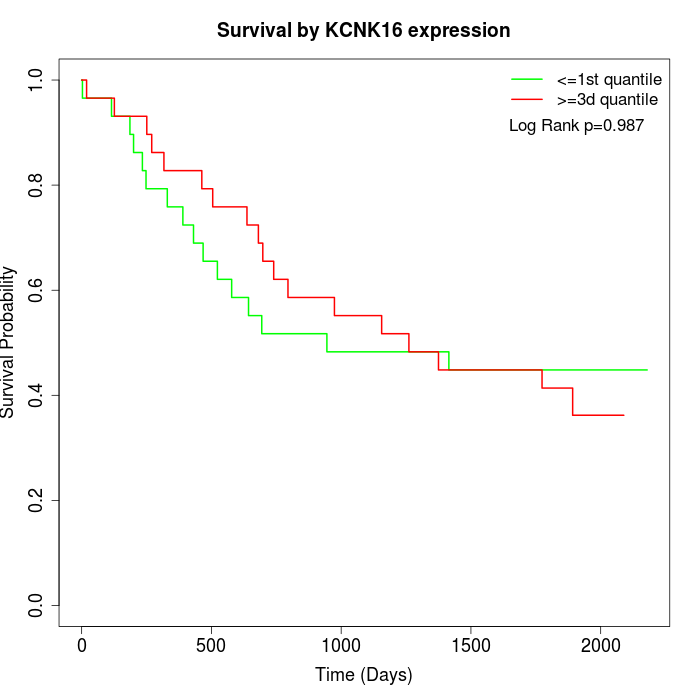
Survival by KCNK16 expression:
Note: Click image to view full size file.
Copy number change of KCNK16:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNK16 | 83795 | 4 | 1 | 25 | |
| GSE20123 | KCNK16 | 83795 | 4 | 1 | 25 | |
| GSE43470 | KCNK16 | 83795 | 6 | 0 | 37 | |
| GSE46452 | KCNK16 | 83795 | 2 | 9 | 48 | |
| GSE47630 | KCNK16 | 83795 | 8 | 4 | 28 | |
| GSE54993 | KCNK16 | 83795 | 3 | 2 | 65 | |
| GSE54994 | KCNK16 | 83795 | 11 | 4 | 38 | |
| GSE60625 | KCNK16 | 83795 | 0 | 1 | 10 | |
| GSE74703 | KCNK16 | 83795 | 6 | 0 | 30 | |
| GSE74704 | KCNK16 | 83795 | 1 | 1 | 18 | |
| TCGA | KCNK16 | 83795 | 17 | 14 | 65 |
Total number of gains: 62; Total number of losses: 37; Total Number of normals: 389.
Somatic mutations of KCNK16:
Generating mutation plots.
Highly correlated genes for KCNK16:
Showing top 20/78 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNK16 | CYP2A13 | 0.710178 | 3 | 0 | 3 |
| KCNK16 | ZNF517 | 0.706755 | 3 | 0 | 3 |
| KCNK16 | CEACAM3 | 0.705677 | 3 | 0 | 3 |
| KCNK16 | LINC00347 | 0.669099 | 3 | 0 | 3 |
| KCNK16 | GDF11 | 0.627369 | 3 | 0 | 3 |
| KCNK16 | CNTD2 | 0.625381 | 5 | 0 | 3 |
| KCNK16 | FBXL18 | 0.624346 | 4 | 0 | 3 |
| KCNK16 | HRH3 | 0.623301 | 4 | 0 | 4 |
| KCNK16 | CA1 | 0.623199 | 3 | 0 | 3 |
| KCNK16 | FGF18 | 0.623017 | 3 | 0 | 3 |
| KCNK16 | LRFN4 | 0.622936 | 3 | 0 | 3 |
| KCNK16 | PKD2L1 | 0.622181 | 3 | 0 | 3 |
| KCNK16 | DDX11-AS1 | 0.621714 | 3 | 0 | 3 |
| KCNK16 | PRAM1 | 0.620916 | 3 | 0 | 3 |
| KCNK16 | ARVCF | 0.615136 | 3 | 0 | 3 |
| KCNK16 | ZFYVE28 | 0.614679 | 3 | 0 | 3 |
| KCNK16 | MOGAT2 | 0.612343 | 4 | 0 | 4 |
| KCNK16 | NANOS3 | 0.611628 | 5 | 0 | 4 |
| KCNK16 | LINC01118 | 0.606798 | 4 | 0 | 3 |
| KCNK16 | ARMC3 | 0.60555 | 3 | 0 | 3 |
For details and further investigation, click here