| Full name: N-acetyltransferase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 8p22 | ||
| Entrez ID: 10 | HGNC ID: HGNC:7646 | Ensembl Gene: ENSG00000156006 | OMIM ID: 612182 |
| Drug and gene relationship at DGIdb | |||
Expression of NAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NAT2 | 10 | 206797_at | 0.0027 | 0.9943 | |
| GSE20347 | NAT2 | 10 | 206797_at | 0.0892 | 0.2440 | |
| GSE23400 | NAT2 | 10 | 206797_at | -0.1987 | 0.0000 | |
| GSE26886 | NAT2 | 10 | 206797_at | -0.1381 | 0.3426 | |
| GSE29001 | NAT2 | 10 | 206797_at | -0.2303 | 0.2287 | |
| GSE38129 | NAT2 | 10 | 206797_at | -0.1313 | 0.0496 | |
| GSE45670 | NAT2 | 10 | 206797_at | 0.0905 | 0.2409 | |
| GSE53622 | NAT2 | 10 | 36227 | -0.3942 | 0.0034 | |
| GSE53624 | NAT2 | 10 | 51354 | -0.1169 | 0.2326 | |
| GSE63941 | NAT2 | 10 | 206797_at | 0.1296 | 0.5143 | |
| GSE77861 | NAT2 | 10 | 206797_at | -0.0030 | 0.9820 | |
| TCGA | NAT2 | 10 | RNAseq | 0.6369 | 0.5819 |
Upregulated datasets: 0; Downregulated datasets: 0.
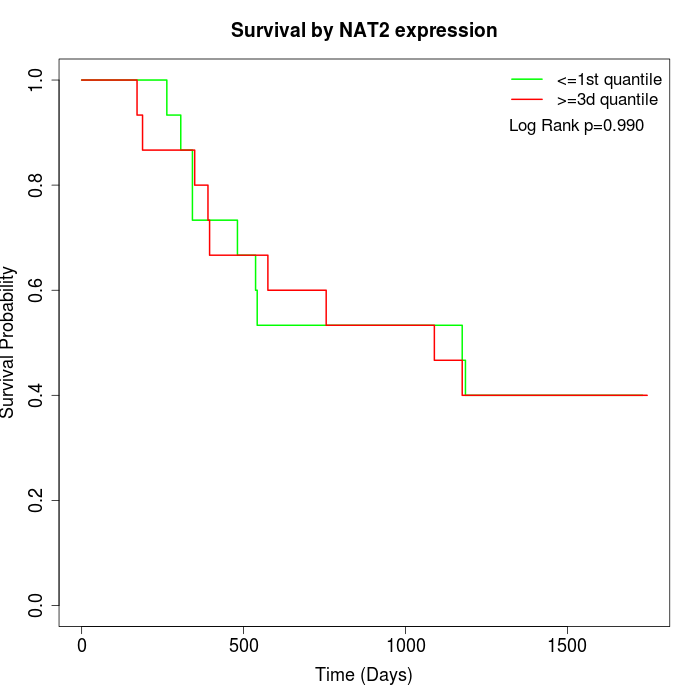
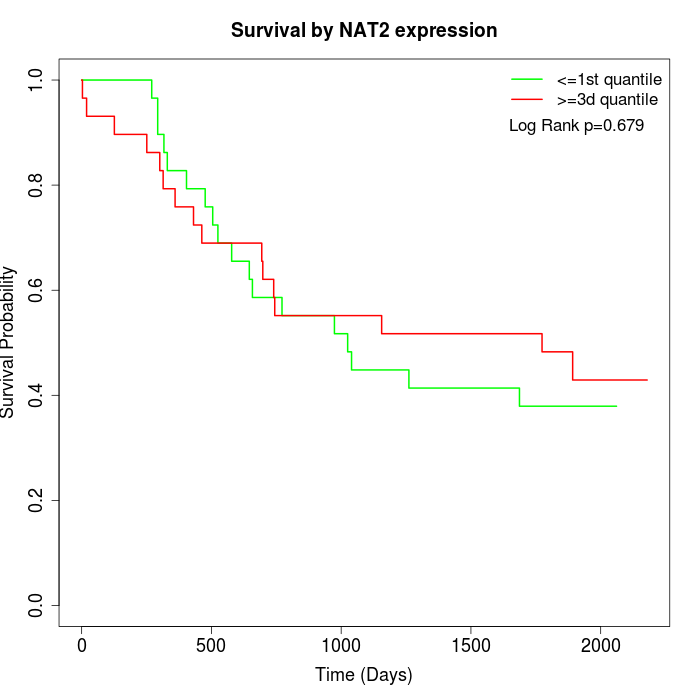
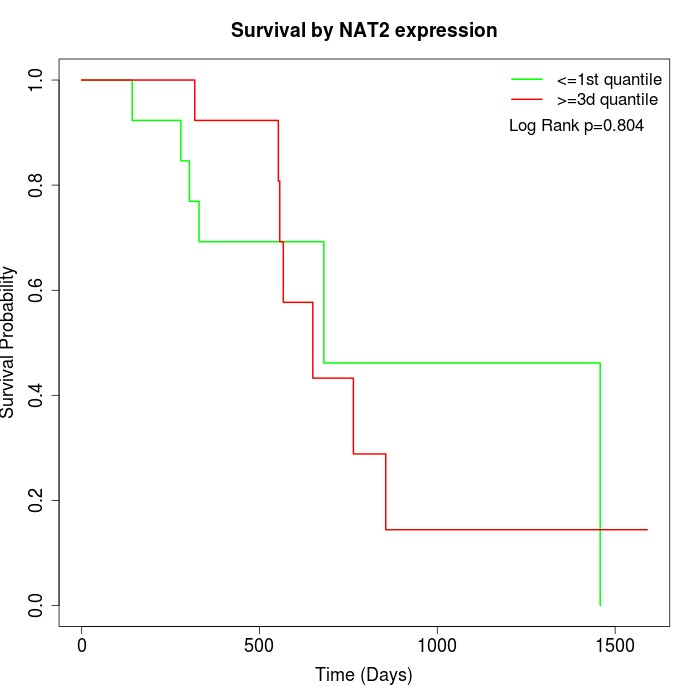
Survival by NAT2 expression:
Note: Click image to view full size file.
Copy number change of NAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NAT2 | 10 | 3 | 10 | 17 | |
| GSE20123 | NAT2 | 10 | 4 | 10 | 16 | |
| GSE43470 | NAT2 | 10 | 4 | 8 | 31 | |
| GSE46452 | NAT2 | 10 | 14 | 13 | 32 | |
| GSE47630 | NAT2 | 10 | 10 | 8 | 22 | |
| GSE54993 | NAT2 | 10 | 2 | 14 | 54 | |
| GSE54994 | NAT2 | 10 | 8 | 18 | 27 | |
| GSE60625 | NAT2 | 10 | 3 | 0 | 8 | |
| GSE74703 | NAT2 | 10 | 4 | 7 | 25 | |
| GSE74704 | NAT2 | 10 | 3 | 7 | 10 | |
| TCGA | NAT2 | 10 | 13 | 42 | 41 |
Total number of gains: 68; Total number of losses: 137; Total Number of normals: 283.
Somatic mutations of NAT2:
Generating mutation plots.
Highly correlated genes for NAT2:
Showing top 20/750 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NAT2 | NOS1 | 0.780736 | 3 | 0 | 3 |
| NAT2 | SIX6 | 0.772479 | 3 | 0 | 3 |
| NAT2 | MLXIPL | 0.7651 | 3 | 0 | 3 |
| NAT2 | ADAMTS12 | 0.722529 | 4 | 0 | 4 |
| NAT2 | IFT122 | 0.717632 | 3 | 0 | 3 |
| NAT2 | RALYL | 0.70955 | 3 | 0 | 3 |
| NAT2 | LGALS14 | 0.705938 | 3 | 0 | 3 |
| NAT2 | GCM1 | 0.705544 | 3 | 0 | 3 |
| NAT2 | SLC45A2 | 0.704324 | 4 | 0 | 4 |
| NAT2 | CDC25C | 0.698946 | 3 | 0 | 3 |
| NAT2 | IFNA10 | 0.698744 | 5 | 0 | 4 |
| NAT2 | CST8 | 0.698539 | 4 | 0 | 3 |
| NAT2 | CRYGC | 0.697218 | 3 | 0 | 3 |
| NAT2 | MATN1 | 0.694226 | 4 | 0 | 3 |
| NAT2 | KIR2DL4 | 0.693837 | 5 | 0 | 5 |
| NAT2 | PRDM11 | 0.692344 | 3 | 0 | 3 |
| NAT2 | MED24 | 0.686232 | 5 | 0 | 4 |
| NAT2 | MEFV | 0.68575 | 3 | 0 | 3 |
| NAT2 | AQP2 | 0.685495 | 4 | 0 | 3 |
| NAT2 | ADM2 | 0.684505 | 5 | 0 | 4 |
For details and further investigation, click here