| Full name: DAB adaptor protein 2 | Alias Symbol: DOC-2 | ||
| Type: protein-coding gene | Cytoband: 5p13.1 | ||
| Entrez ID: 1601 | HGNC ID: HGNC:2662 | Ensembl Gene: ENSG00000153071 | OMIM ID: 601236 |
| Drug and gene relationship at DGIdb | |||
Expression of DAB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DAB2 | 1601 | 201280_s_at | -0.3246 | 0.7345 | |
| GSE20347 | DAB2 | 1601 | 201280_s_at | 0.9871 | 0.0012 | |
| GSE23400 | DAB2 | 1601 | 210757_x_at | 0.2640 | 0.0277 | |
| GSE26886 | DAB2 | 1601 | 201278_at | 1.2430 | 0.0096 | |
| GSE29001 | DAB2 | 1601 | 210757_x_at | 0.7757 | 0.0024 | |
| GSE38129 | DAB2 | 1601 | 201280_s_at | 0.3936 | 0.1459 | |
| GSE45670 | DAB2 | 1601 | 201278_at | -1.4935 | 0.0009 | |
| GSE53622 | DAB2 | 1601 | 45948 | -0.1291 | 0.3920 | |
| GSE53624 | DAB2 | 1601 | 45948 | 0.2454 | 0.0479 | |
| GSE63941 | DAB2 | 1601 | 210757_x_at | -5.5005 | 0.0005 | |
| GSE77861 | DAB2 | 1601 | 210757_x_at | 0.5831 | 0.1888 | |
| GSE97050 | DAB2 | 1601 | A_33_P3338928 | -0.1292 | 0.7392 | |
| SRP007169 | DAB2 | 1601 | RNAseq | 2.6186 | 0.0000 | |
| SRP008496 | DAB2 | 1601 | RNAseq | 2.7907 | 0.0000 | |
| SRP064894 | DAB2 | 1601 | RNAseq | 0.7042 | 0.0410 | |
| SRP133303 | DAB2 | 1601 | RNAseq | 0.3367 | 0.2832 | |
| SRP159526 | DAB2 | 1601 | RNAseq | 0.9829 | 0.0005 | |
| SRP193095 | DAB2 | 1601 | RNAseq | 1.1387 | 0.0000 | |
| SRP219564 | DAB2 | 1601 | RNAseq | 0.9458 | 0.1250 | |
| TCGA | DAB2 | 1601 | RNAseq | 0.0374 | 0.7151 |
Upregulated datasets: 4; Downregulated datasets: 2.
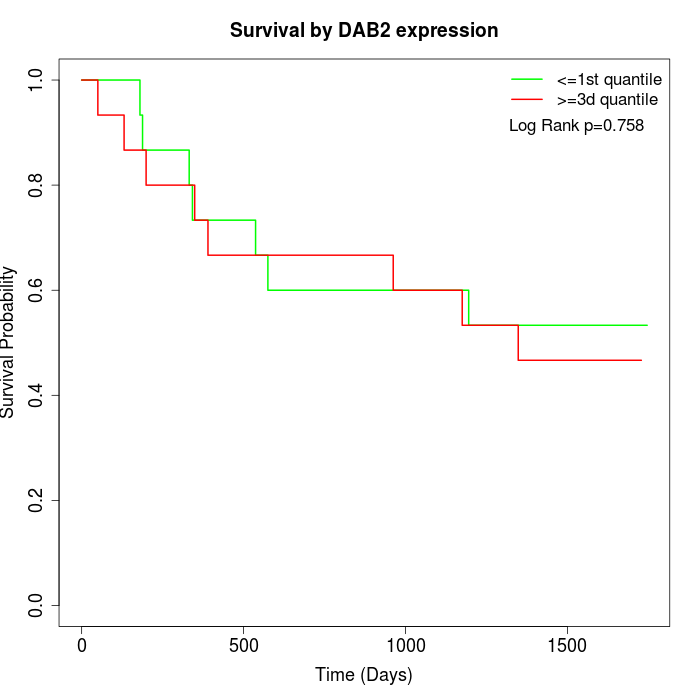
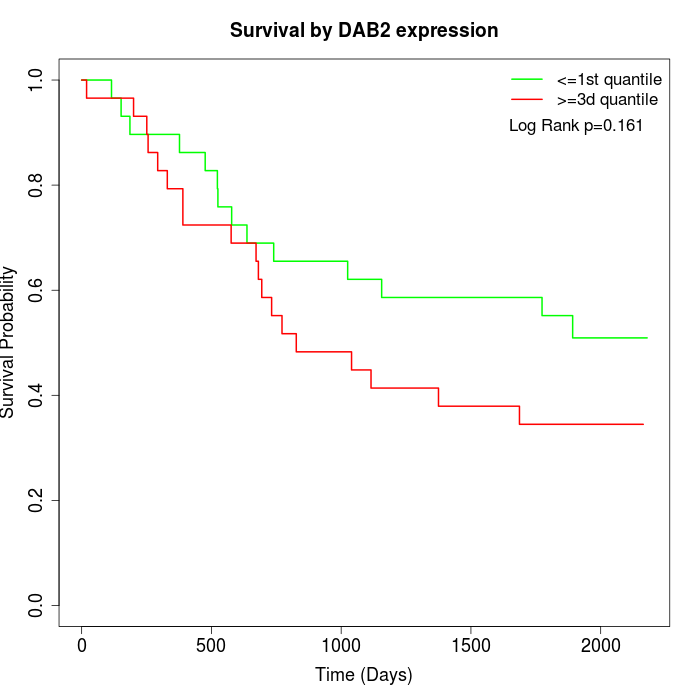
Survival by DAB2 expression:
Note: Click image to view full size file.
Copy number change of DAB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DAB2 | 1601 | 10 | 0 | 20 | |
| GSE20123 | DAB2 | 1601 | 10 | 0 | 20 | |
| GSE43470 | DAB2 | 1601 | 17 | 0 | 26 | |
| GSE46452 | DAB2 | 1601 | 5 | 22 | 32 | |
| GSE47630 | DAB2 | 1601 | 6 | 12 | 22 | |
| GSE54993 | DAB2 | 1601 | 5 | 4 | 61 | |
| GSE54994 | DAB2 | 1601 | 24 | 2 | 27 | |
| GSE60625 | DAB2 | 1601 | 0 | 0 | 11 | |
| GSE74703 | DAB2 | 1601 | 13 | 0 | 23 | |
| GSE74704 | DAB2 | 1601 | 8 | 0 | 12 | |
| TCGA | DAB2 | 1601 | 52 | 6 | 38 |
Total number of gains: 150; Total number of losses: 46; Total Number of normals: 292.
Somatic mutations of DAB2:
Generating mutation plots.
Highly correlated genes for DAB2:
Showing top 20/1043 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DAB2 | BICC1 | 0.807367 | 9 | 0 | 9 |
| DAB2 | FBN1 | 0.780719 | 11 | 0 | 11 |
| DAB2 | PMP22 | 0.765371 | 12 | 0 | 11 |
| DAB2 | CCDC80 | 0.764038 | 8 | 0 | 7 |
| DAB2 | GLIPR2 | 0.74661 | 8 | 0 | 8 |
| DAB2 | CNRIP1 | 0.745855 | 9 | 0 | 8 |
| DAB2 | OLFML3 | 0.737919 | 13 | 0 | 13 |
| DAB2 | EFEMP2 | 0.736293 | 12 | 0 | 11 |
| DAB2 | MOB3A | 0.733256 | 4 | 0 | 3 |
| DAB2 | LINC01279 | 0.730668 | 5 | 0 | 4 |
| DAB2 | RARRES2 | 0.726675 | 11 | 0 | 10 |
| DAB2 | LAMA4 | 0.726468 | 12 | 0 | 12 |
| DAB2 | FSTL1 | 0.721931 | 13 | 0 | 13 |
| DAB2 | C11orf96 | 0.72052 | 5 | 0 | 5 |
| DAB2 | TSPAN4 | 0.717991 | 12 | 0 | 11 |
| DAB2 | COLEC12 | 0.716409 | 12 | 0 | 12 |
| DAB2 | BHLHE22 | 0.715553 | 6 | 0 | 6 |
| DAB2 | F13A1 | 0.71479 | 9 | 0 | 8 |
| DAB2 | OLFML1 | 0.712894 | 13 | 0 | 13 |
| DAB2 | DDR2 | 0.711809 | 12 | 0 | 11 |
For details and further investigation, click here