| Full name: dual adaptor of phosphotyrosine and 3-phosphoinositides 1 | Alias Symbol: BAM32 | ||
| Type: protein-coding gene | Cytoband: 4q23 | ||
| Entrez ID: 27071 | HGNC ID: HGNC:16500 | Ensembl Gene: ENSG00000070190 | OMIM ID: 605768 |
| Drug and gene relationship at DGIdb | |||
DAPP1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04662 | B cell receptor signaling pathway |
Expression of DAPP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DAPP1 | 27071 | 219290_x_at | -0.2434 | 0.8197 | |
| GSE20347 | DAPP1 | 27071 | 219290_x_at | -0.2908 | 0.1566 | |
| GSE23400 | DAPP1 | 27071 | 219290_x_at | -0.2986 | 0.0000 | |
| GSE26886 | DAPP1 | 27071 | 219290_x_at | -1.2480 | 0.0000 | |
| GSE29001 | DAPP1 | 27071 | 219290_x_at | -0.9335 | 0.0052 | |
| GSE38129 | DAPP1 | 27071 | 219290_x_at | -0.3881 | 0.1634 | |
| GSE45670 | DAPP1 | 27071 | 219290_x_at | -0.0399 | 0.8566 | |
| GSE53622 | DAPP1 | 27071 | 21884 | -1.2372 | 0.0000 | |
| GSE53624 | DAPP1 | 27071 | 21884 | -2.0164 | 0.0000 | |
| GSE63941 | DAPP1 | 27071 | 219290_x_at | 0.6151 | 0.2587 | |
| GSE77861 | DAPP1 | 27071 | 219290_x_at | -0.3363 | 0.3559 | |
| GSE97050 | DAPP1 | 27071 | A_23_P255444 | -0.0811 | 0.9335 | |
| SRP007169 | DAPP1 | 27071 | RNAseq | -1.3839 | 0.0025 | |
| SRP008496 | DAPP1 | 27071 | RNAseq | -1.9275 | 0.0000 | |
| SRP064894 | DAPP1 | 27071 | RNAseq | -2.4402 | 0.0000 | |
| SRP133303 | DAPP1 | 27071 | RNAseq | -1.2344 | 0.0000 | |
| SRP159526 | DAPP1 | 27071 | RNAseq | -1.9288 | 0.0030 | |
| SRP193095 | DAPP1 | 27071 | RNAseq | -1.3452 | 0.0000 | |
| SRP219564 | DAPP1 | 27071 | RNAseq | -2.8295 | 0.0020 | |
| TCGA | DAPP1 | 27071 | RNAseq | 0.6147 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 10.
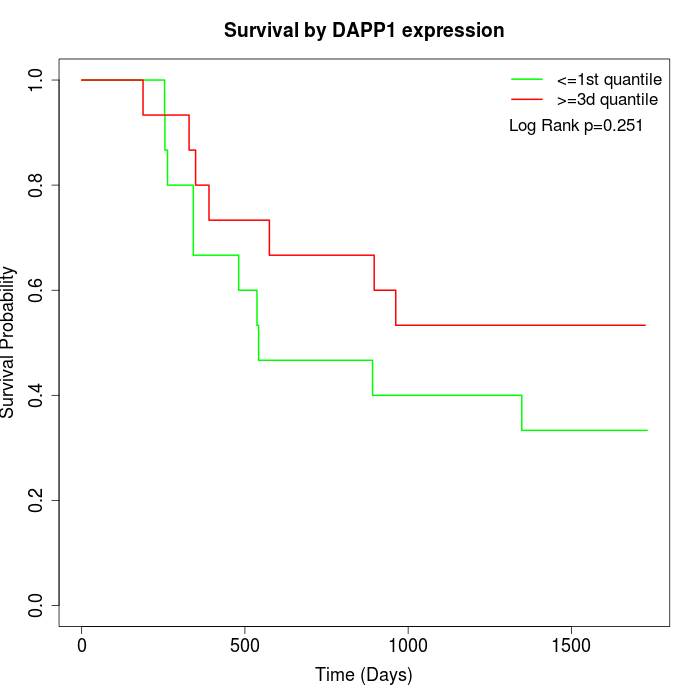
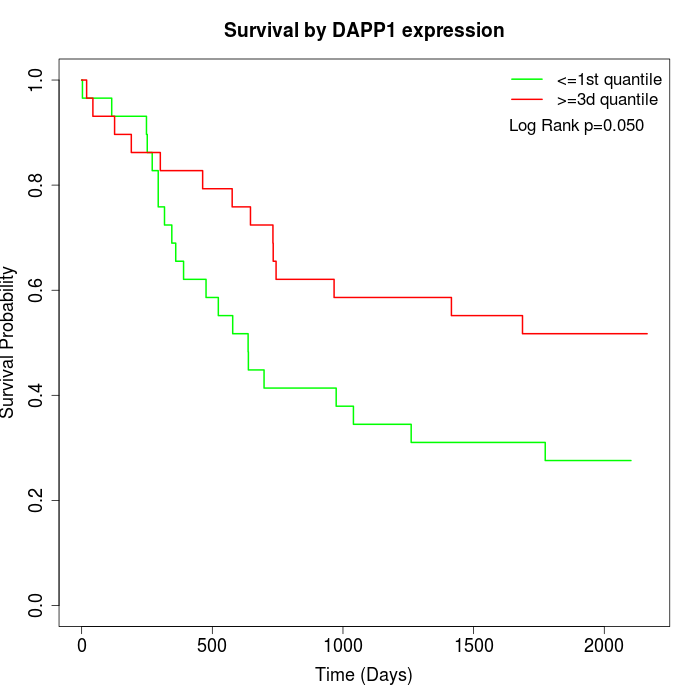
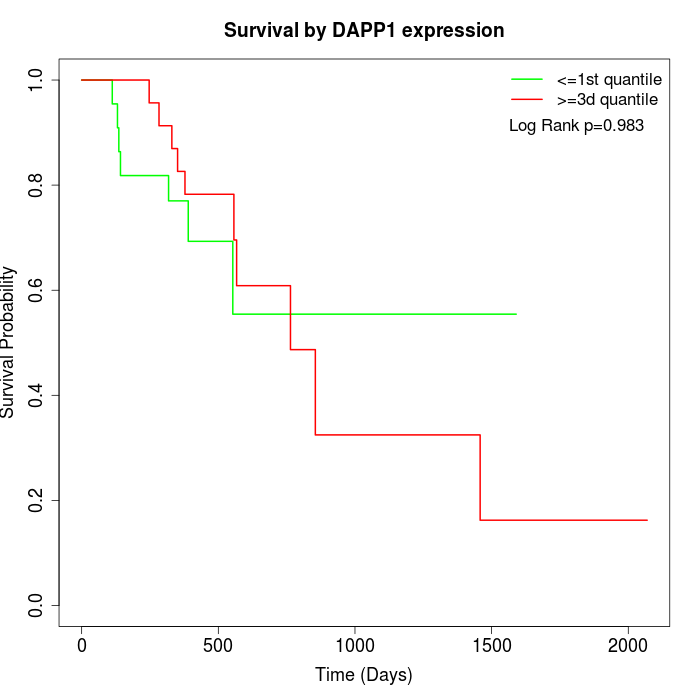
Survival by DAPP1 expression:
Note: Click image to view full size file.
Copy number change of DAPP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DAPP1 | 27071 | 0 | 12 | 18 | |
| GSE20123 | DAPP1 | 27071 | 0 | 12 | 18 | |
| GSE43470 | DAPP1 | 27071 | 0 | 13 | 30 | |
| GSE46452 | DAPP1 | 27071 | 1 | 36 | 22 | |
| GSE47630 | DAPP1 | 27071 | 0 | 20 | 20 | |
| GSE54993 | DAPP1 | 27071 | 9 | 0 | 61 | |
| GSE54994 | DAPP1 | 27071 | 1 | 11 | 41 | |
| GSE60625 | DAPP1 | 27071 | 0 | 3 | 8 | |
| GSE74703 | DAPP1 | 27071 | 0 | 11 | 25 | |
| GSE74704 | DAPP1 | 27071 | 0 | 6 | 14 | |
| TCGA | DAPP1 | 27071 | 8 | 40 | 48 |
Total number of gains: 19; Total number of losses: 164; Total Number of normals: 305.
Somatic mutations of DAPP1:
Generating mutation plots.
Highly correlated genes for DAPP1:
Showing top 20/1108 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DAPP1 | NUAK2 | 0.833374 | 3 | 0 | 3 |
| DAPP1 | EPGN | 0.825917 | 3 | 0 | 3 |
| DAPP1 | ELOVL1 | 0.793262 | 3 | 0 | 3 |
| DAPP1 | KIF21A | 0.772143 | 4 | 0 | 4 |
| DAPP1 | ARAP2 | 0.769725 | 3 | 0 | 3 |
| DAPP1 | ATP13A4 | 0.761451 | 5 | 0 | 5 |
| DAPP1 | CCNYL1 | 0.761344 | 4 | 0 | 4 |
| DAPP1 | MPZL3 | 0.760249 | 7 | 0 | 6 |
| DAPP1 | DUOXA1 | 0.752326 | 6 | 0 | 6 |
| DAPP1 | ZNF431 | 0.751211 | 5 | 0 | 5 |
| DAPP1 | S100A16 | 0.747753 | 6 | 0 | 5 |
| DAPP1 | KCNK6 | 0.74536 | 6 | 0 | 6 |
| DAPP1 | ZDHHC5 | 0.744641 | 5 | 0 | 5 |
| DAPP1 | BTBD11 | 0.743315 | 6 | 0 | 6 |
| DAPP1 | TMEM45B | 0.743297 | 6 | 0 | 6 |
| DAPP1 | ANKRD31 | 0.738651 | 4 | 0 | 4 |
| DAPP1 | CKMT1A | 0.738122 | 3 | 0 | 3 |
| DAPP1 | DENND2C | 0.72999 | 7 | 0 | 6 |
| DAPP1 | C15orf48 | 0.72909 | 7 | 0 | 7 |
| DAPP1 | LTA4H | 0.728568 | 3 | 0 | 3 |
For details and further investigation, click here