| Full name: decapping mRNA 2 | Alias Symbol: NUDT20 | ||
| Type: protein-coding gene | Cytoband: 5q22.2 | ||
| Entrez ID: 167227 | HGNC ID: HGNC:24452 | Ensembl Gene: ENSG00000172795 | OMIM ID: 609844 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of DCP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCP2 | 167227 | 212919_at | 0.5221 | 0.2143 | |
| GSE20347 | DCP2 | 167227 | 212919_at | 0.0171 | 0.9215 | |
| GSE23400 | DCP2 | 167227 | 212919_at | 0.2541 | 0.0002 | |
| GSE26886 | DCP2 | 167227 | 212919_at | 0.0162 | 0.9574 | |
| GSE29001 | DCP2 | 167227 | 212919_at | 0.1208 | 0.6962 | |
| GSE38129 | DCP2 | 167227 | 212919_at | 0.0663 | 0.7127 | |
| GSE45670 | DCP2 | 167227 | 212919_at | 0.1401 | 0.4675 | |
| GSE53622 | DCP2 | 167227 | 47678 | 0.1689 | 0.1429 | |
| GSE53624 | DCP2 | 167227 | 47678 | 0.0410 | 0.7083 | |
| GSE63941 | DCP2 | 167227 | 212919_at | -0.2254 | 0.7031 | |
| GSE77861 | DCP2 | 167227 | 212919_at | -0.1240 | 0.7314 | |
| GSE97050 | DCP2 | 167227 | A_24_P886040 | 0.2004 | 0.3436 | |
| SRP007169 | DCP2 | 167227 | RNAseq | 0.0004 | 0.9991 | |
| SRP008496 | DCP2 | 167227 | RNAseq | 0.0118 | 0.9537 | |
| SRP064894 | DCP2 | 167227 | RNAseq | 0.3143 | 0.0911 | |
| SRP133303 | DCP2 | 167227 | RNAseq | 0.2489 | 0.1352 | |
| SRP159526 | DCP2 | 167227 | RNAseq | -0.1890 | 0.4034 | |
| SRP193095 | DCP2 | 167227 | RNAseq | -0.2247 | 0.0146 | |
| SRP219564 | DCP2 | 167227 | RNAseq | -0.0776 | 0.8095 | |
| TCGA | DCP2 | 167227 | RNAseq | 0.0537 | 0.3280 |
Upregulated datasets: 0; Downregulated datasets: 0.
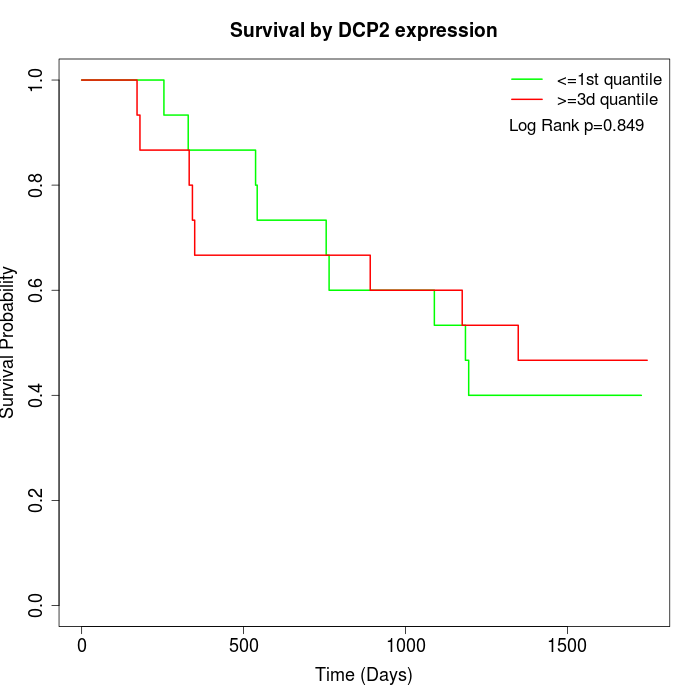
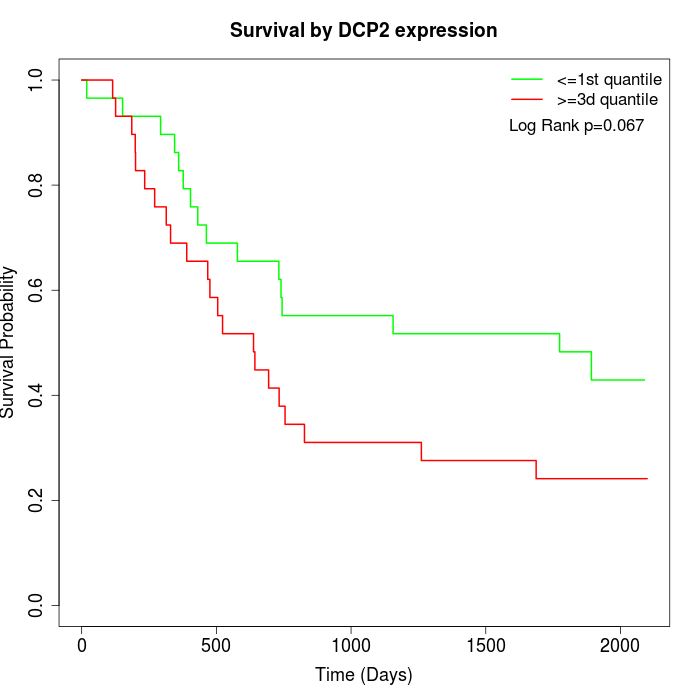
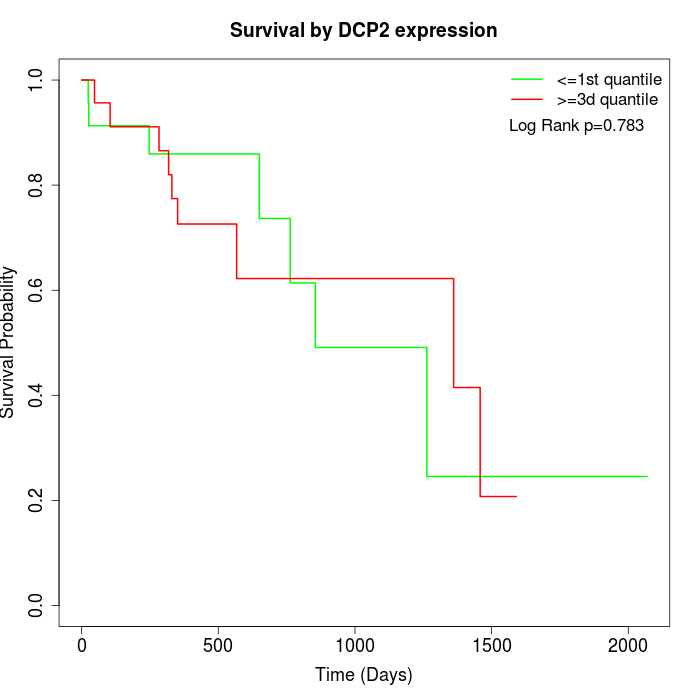
Survival by DCP2 expression:
Note: Click image to view full size file.
Copy number change of DCP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCP2 | 167227 | 1 | 13 | 16 | |
| GSE20123 | DCP2 | 167227 | 1 | 13 | 16 | |
| GSE43470 | DCP2 | 167227 | 3 | 8 | 32 | |
| GSE46452 | DCP2 | 167227 | 0 | 28 | 31 | |
| GSE47630 | DCP2 | 167227 | 0 | 21 | 19 | |
| GSE54993 | DCP2 | 167227 | 9 | 1 | 60 | |
| GSE54994 | DCP2 | 167227 | 1 | 17 | 35 | |
| GSE60625 | DCP2 | 167227 | 0 | 0 | 11 | |
| GSE74703 | DCP2 | 167227 | 2 | 6 | 28 | |
| GSE74704 | DCP2 | 167227 | 1 | 7 | 12 | |
| TCGA | DCP2 | 167227 | 2 | 41 | 53 |
Total number of gains: 20; Total number of losses: 155; Total Number of normals: 313.
Somatic mutations of DCP2:
Generating mutation plots.
Highly correlated genes for DCP2:
Showing top 20/399 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCP2 | PSEN1 | 0.818096 | 3 | 0 | 3 |
| DCP2 | NAF1 | 0.750882 | 3 | 0 | 3 |
| DCP2 | ARL10 | 0.750338 | 3 | 0 | 3 |
| DCP2 | CHCHD7 | 0.739306 | 3 | 0 | 3 |
| DCP2 | SOS2 | 0.732735 | 3 | 0 | 3 |
| DCP2 | ZNF830 | 0.731754 | 3 | 0 | 3 |
| DCP2 | SRRD | 0.725893 | 3 | 0 | 3 |
| DCP2 | NAA30 | 0.71968 | 3 | 0 | 3 |
| DCP2 | EAF1 | 0.707953 | 3 | 0 | 3 |
| DCP2 | WDR12 | 0.707573 | 3 | 0 | 3 |
| DCP2 | TRUB1 | 0.704382 | 4 | 0 | 4 |
| DCP2 | EIF2S2 | 0.701536 | 3 | 0 | 3 |
| DCP2 | STYX | 0.700675 | 3 | 0 | 3 |
| DCP2 | NUDT3 | 0.698485 | 3 | 0 | 3 |
| DCP2 | POLR3GL | 0.692227 | 3 | 0 | 3 |
| DCP2 | ISCA2 | 0.691216 | 3 | 0 | 3 |
| DCP2 | SNX12 | 0.690098 | 3 | 0 | 3 |
| DCP2 | ATG4C | 0.689786 | 3 | 0 | 3 |
| DCP2 | PHOSPHO2 | 0.687592 | 3 | 0 | 3 |
| DCP2 | GZF1 | 0.685121 | 3 | 0 | 3 |
For details and further investigation, click here