| Full name: dopachrome tautomerase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 13q32.1 | ||
| Entrez ID: 1638 | HGNC ID: HGNC:2709 | Ensembl Gene: ENSG00000080166 | OMIM ID: 191275 |
| Drug and gene relationship at DGIdb | |||
DCT involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04916 | Melanogenesis |
Expression of DCT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCT | 1638 | 216512_s_at | -0.1588 | 0.4992 | |
| GSE20347 | DCT | 1638 | 205338_s_at | 0.1984 | 0.2136 | |
| GSE23400 | DCT | 1638 | 205338_s_at | -0.2409 | 0.0000 | |
| GSE26886 | DCT | 1638 | 216512_s_at | -0.0691 | 0.6333 | |
| GSE29001 | DCT | 1638 | 205338_s_at | -0.5238 | 0.1300 | |
| GSE38129 | DCT | 1638 | 205338_s_at | -0.0976 | 0.4955 | |
| GSE45670 | DCT | 1638 | 216512_s_at | 0.0793 | 0.2928 | |
| GSE53622 | DCT | 1638 | 112373 | -0.7857 | 0.0000 | |
| GSE53624 | DCT | 1638 | 112373 | -1.0142 | 0.0000 | |
| GSE63941 | DCT | 1638 | 216512_s_at | 0.0712 | 0.7228 | |
| GSE77861 | DCT | 1638 | 216512_s_at | -0.2172 | 0.1684 | |
| TCGA | DCT | 1638 | RNAseq | -0.2271 | 0.8387 |
Upregulated datasets: 0; Downregulated datasets: 1.
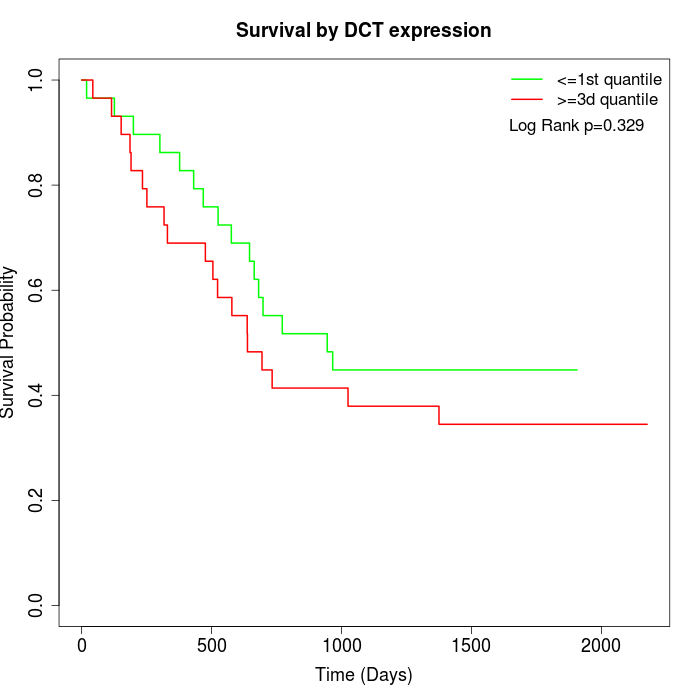
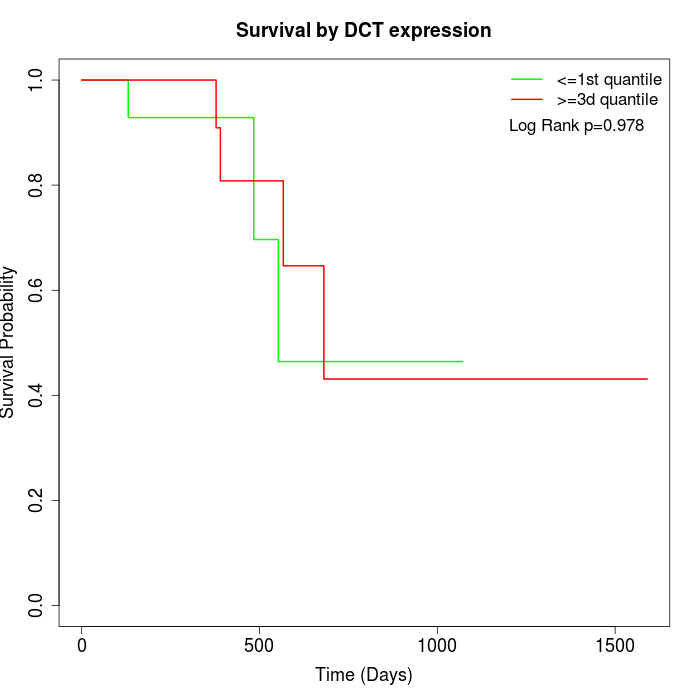
Survival by DCT expression:
Note: Click image to view full size file.
Copy number change of DCT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCT | 1638 | 4 | 9 | 17 | |
| GSE20123 | DCT | 1638 | 4 | 8 | 18 | |
| GSE43470 | DCT | 1638 | 2 | 12 | 29 | |
| GSE46452 | DCT | 1638 | 0 | 32 | 27 | |
| GSE47630 | DCT | 1638 | 3 | 27 | 10 | |
| GSE54993 | DCT | 1638 | 11 | 3 | 56 | |
| GSE54994 | DCT | 1638 | 3 | 14 | 36 | |
| GSE60625 | DCT | 1638 | 0 | 3 | 8 | |
| GSE74703 | DCT | 1638 | 2 | 10 | 24 | |
| GSE74704 | DCT | 1638 | 2 | 8 | 10 | |
| TCGA | DCT | 1638 | 14 | 33 | 49 |
Total number of gains: 45; Total number of losses: 159; Total Number of normals: 284.
Somatic mutations of DCT:
Generating mutation plots.
Highly correlated genes for DCT:
Showing top 20/156 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCT | MYL4 | 0.714446 | 3 | 0 | 3 |
| DCT | CD1E | 0.709681 | 3 | 0 | 3 |
| DCT | CACNA1A | 0.643562 | 3 | 0 | 3 |
| DCT | PAX8 | 0.640557 | 4 | 0 | 3 |
| DCT | LTC4S | 0.637318 | 3 | 0 | 3 |
| DCT | AGRP | 0.624353 | 5 | 0 | 3 |
| DCT | C9orf116 | 0.623873 | 3 | 0 | 3 |
| DCT | HCRT | 0.62373 | 3 | 0 | 3 |
| DCT | TBC1D21 | 0.623547 | 4 | 0 | 4 |
| DCT | CA8 | 0.622396 | 5 | 0 | 4 |
| DCT | FGF16 | 0.62232 | 3 | 0 | 3 |
| DCT | APOB | 0.618455 | 4 | 0 | 4 |
| DCT | ZNF853 | 0.617825 | 3 | 0 | 3 |
| DCT | NPY2R | 0.614092 | 3 | 0 | 3 |
| DCT | GALR3 | 0.612956 | 4 | 0 | 3 |
| DCT | FUT7 | 0.612809 | 3 | 0 | 3 |
| DCT | OMP | 0.611982 | 3 | 0 | 3 |
| DCT | SEMA3B | 0.610282 | 3 | 0 | 3 |
| DCT | CYP11B2 | 0.608346 | 3 | 0 | 3 |
| DCT | ALDH3B1 | 0.605549 | 4 | 0 | 3 |
For details and further investigation, click here