| Full name: distal-less homeobox 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q31.1 | ||
| Entrez ID: 1745 | HGNC ID: HGNC:2914 | Ensembl Gene: ENSG00000144355 | OMIM ID: 600029 |
| Drug and gene relationship at DGIdb | |||
Expression of DLX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DLX1 | 1745 | 1560100_at | -0.0716 | 0.8402 | |
| GSE26886 | DLX1 | 1745 | 1560100_at | -0.0119 | 0.9270 | |
| GSE45670 | DLX1 | 1745 | 1560100_at | 0.0119 | 0.9224 | |
| GSE53622 | DLX1 | 1745 | 24136 | 2.5024 | 0.0000 | |
| GSE53624 | DLX1 | 1745 | 24136 | 3.6037 | 0.0000 | |
| GSE63941 | DLX1 | 1745 | 242138_at | 2.0159 | 0.0472 | |
| GSE77861 | DLX1 | 1745 | 1560100_at | -0.1625 | 0.0813 | |
| GSE97050 | DLX1 | 1745 | A_32_P142818 | 0.2459 | 0.4587 | |
| SRP159526 | DLX1 | 1745 | RNAseq | 5.0596 | 0.0000 | |
| TCGA | DLX1 | 1745 | RNAseq | 1.7897 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
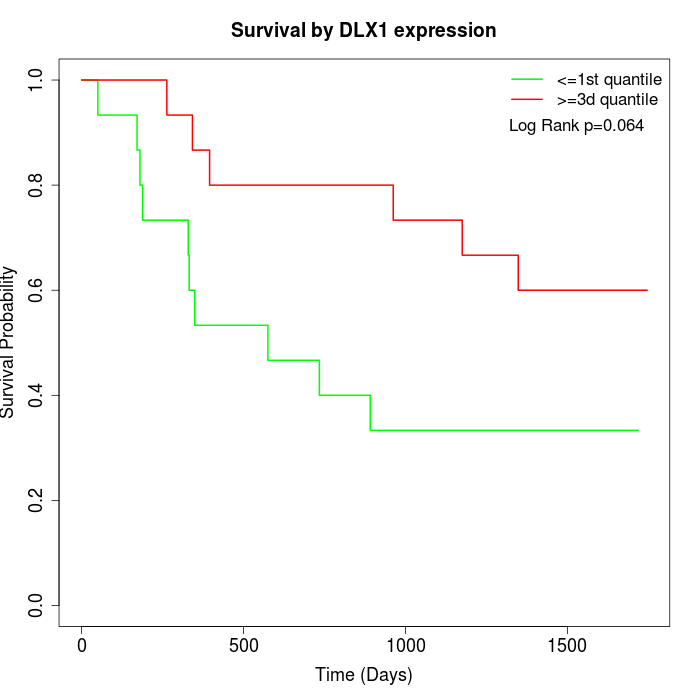
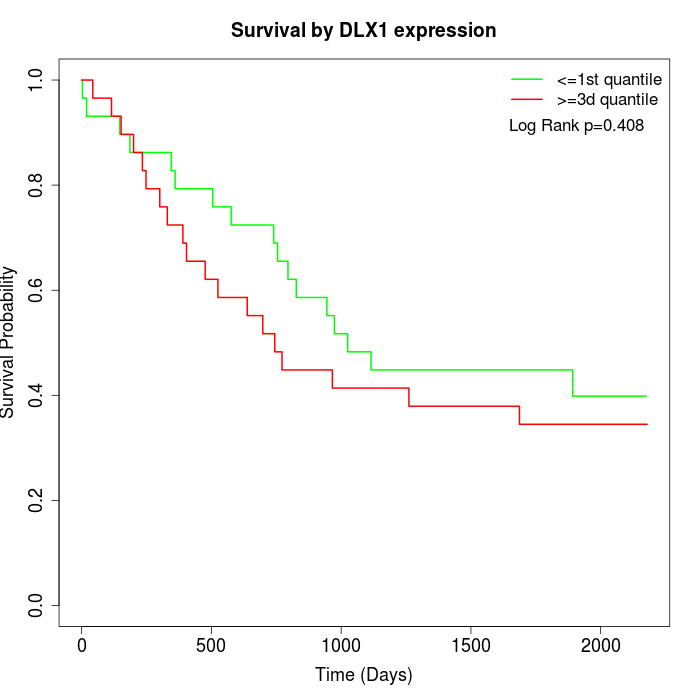
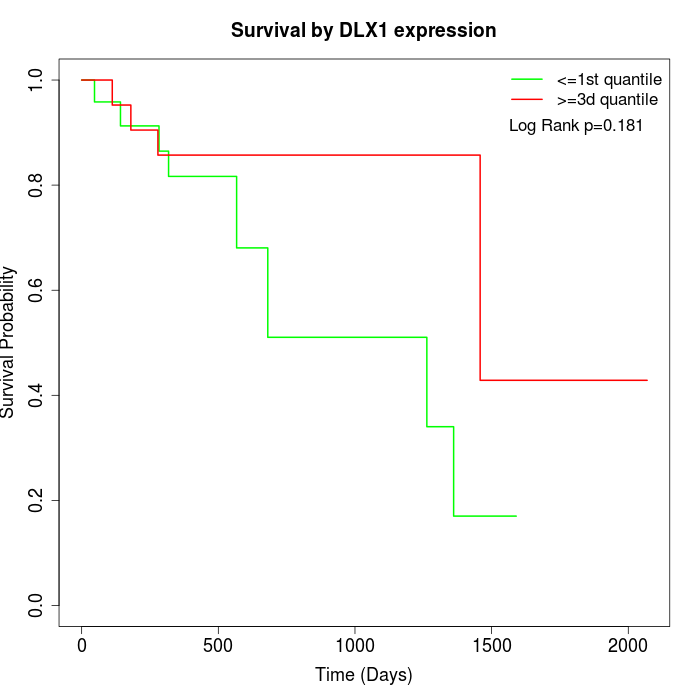
Survival by DLX1 expression:
Note: Click image to view full size file.
Copy number change of DLX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DLX1 | 1745 | 9 | 0 | 21 | |
| GSE20123 | DLX1 | 1745 | 9 | 0 | 21 | |
| GSE43470 | DLX1 | 1745 | 6 | 1 | 36 | |
| GSE46452 | DLX1 | 1745 | 1 | 4 | 54 | |
| GSE47630 | DLX1 | 1745 | 5 | 3 | 32 | |
| GSE54993 | DLX1 | 1745 | 0 | 5 | 65 | |
| GSE54994 | DLX1 | 1745 | 11 | 3 | 39 | |
| GSE60625 | DLX1 | 1745 | 0 | 3 | 8 | |
| GSE74703 | DLX1 | 1745 | 5 | 1 | 30 | |
| GSE74704 | DLX1 | 1745 | 5 | 0 | 15 | |
| TCGA | DLX1 | 1745 | 28 | 6 | 62 |
Total number of gains: 79; Total number of losses: 26; Total Number of normals: 383.
Somatic mutations of DLX1:
Generating mutation plots.
Highly correlated genes for DLX1:
Showing top 20/83 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DLX1 | MCM4 | 0.673466 | 3 | 0 | 3 |
| DLX1 | MMP11 | 0.669889 | 3 | 0 | 3 |
| DLX1 | YEATS2 | 0.665021 | 3 | 0 | 3 |
| DLX1 | FKBP9 | 0.661763 | 3 | 0 | 3 |
| DLX1 | ACYP1 | 0.656484 | 3 | 0 | 3 |
| DLX1 | CHRNA1 | 0.640573 | 3 | 0 | 3 |
| DLX1 | MATN3 | 0.640519 | 3 | 0 | 3 |
| DLX1 | FRMPD3 | 0.623677 | 3 | 0 | 3 |
| DLX1 | MAD2L2 | 0.621522 | 3 | 0 | 3 |
| DLX1 | SLC19A1 | 0.618304 | 3 | 0 | 3 |
| DLX1 | CHPF | 0.608487 | 3 | 0 | 3 |
| DLX1 | PMEPA1 | 0.607695 | 3 | 0 | 3 |
| DLX1 | BCL7A | 0.605486 | 3 | 0 | 3 |
| DLX1 | KIF19 | 0.604087 | 3 | 0 | 3 |
| DLX1 | CHRNA9 | 0.603437 | 3 | 0 | 3 |
| DLX1 | MMP28 | 0.599641 | 3 | 0 | 3 |
| DLX1 | NME4 | 0.599191 | 3 | 0 | 3 |
| DLX1 | DLX2 | 0.597378 | 6 | 0 | 3 |
| DLX1 | GRTP1-AS1 | 0.596746 | 3 | 0 | 3 |
| DLX1 | FAM71D | 0.594321 | 5 | 0 | 5 |
For details and further investigation, click here