| Full name: protein kinase AMP-activated non-catalytic subunit gamma 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.12 | ||
| Entrez ID: 5571 | HGNC ID: HGNC:9385 | Ensembl Gene: ENSG00000181929 | OMIM ID: 602742 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PRKAG1 involved pathways:
Expression of PRKAG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRKAG1 | 5571 | 201805_at | -0.4129 | 0.2863 | |
| GSE20347 | PRKAG1 | 5571 | 201805_at | -0.2353 | 0.0410 | |
| GSE23400 | PRKAG1 | 5571 | 201805_at | -0.1130 | 0.0322 | |
| GSE26886 | PRKAG1 | 5571 | 201805_at | -0.9610 | 0.0000 | |
| GSE29001 | PRKAG1 | 5571 | 201805_at | -0.2573 | 0.2219 | |
| GSE38129 | PRKAG1 | 5571 | 201805_at | -0.1650 | 0.1627 | |
| GSE45670 | PRKAG1 | 5571 | 201805_at | -0.0476 | 0.7168 | |
| GSE53622 | PRKAG1 | 5571 | 28628 | -0.1377 | 0.0195 | |
| GSE53624 | PRKAG1 | 5571 | 28628 | -0.2883 | 0.0000 | |
| GSE63941 | PRKAG1 | 5571 | 201805_at | 1.1119 | 0.0042 | |
| GSE77861 | PRKAG1 | 5571 | 201805_at | -0.1119 | 0.4766 | |
| GSE97050 | PRKAG1 | 5571 | A_23_P36513 | -0.4532 | 0.1324 | |
| SRP007169 | PRKAG1 | 5571 | RNAseq | -0.1084 | 0.7493 | |
| SRP008496 | PRKAG1 | 5571 | RNAseq | -0.1981 | 0.4030 | |
| SRP064894 | PRKAG1 | 5571 | RNAseq | -0.3355 | 0.0013 | |
| SRP133303 | PRKAG1 | 5571 | RNAseq | -0.0350 | 0.7530 | |
| SRP159526 | PRKAG1 | 5571 | RNAseq | -0.4111 | 0.0537 | |
| SRP193095 | PRKAG1 | 5571 | RNAseq | -0.4186 | 0.0002 | |
| SRP219564 | PRKAG1 | 5571 | RNAseq | -0.4287 | 0.0637 | |
| TCGA | PRKAG1 | 5571 | RNAseq | -0.0036 | 0.9436 |
Upregulated datasets: 1; Downregulated datasets: 0.
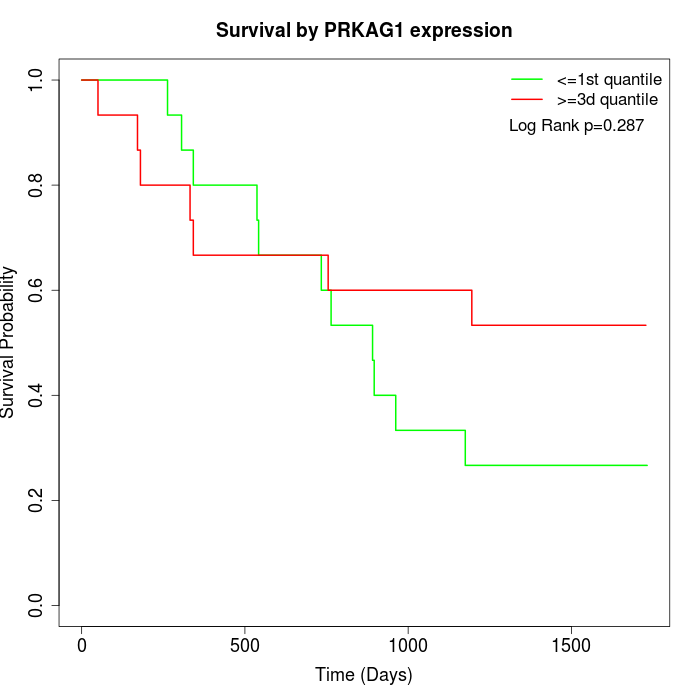
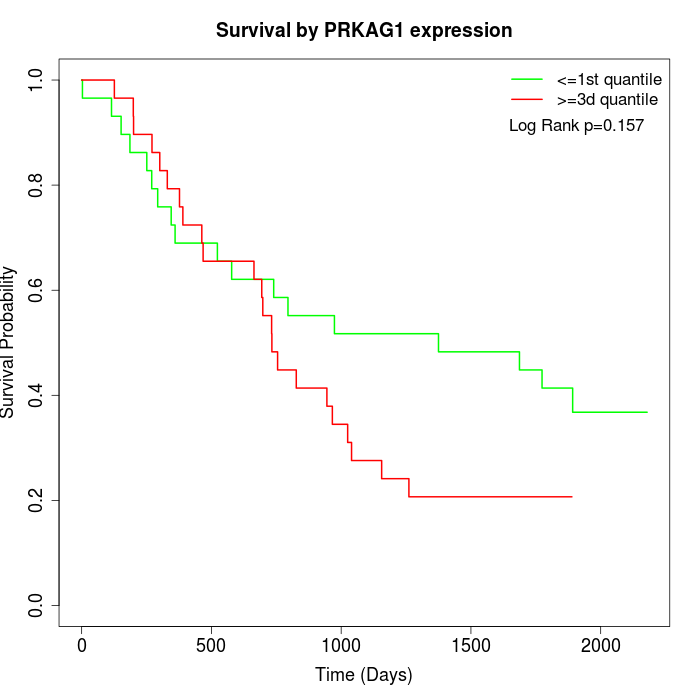
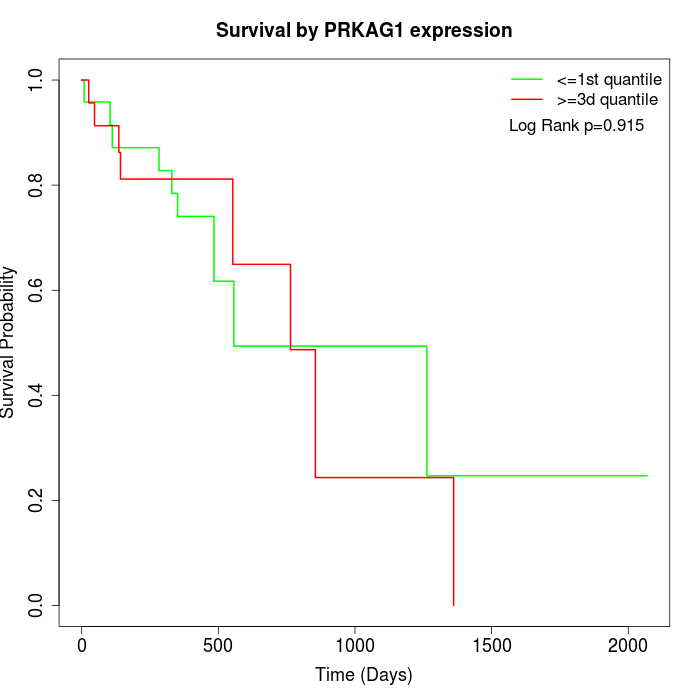
Survival by PRKAG1 expression:
Note: Click image to view full size file.
Copy number change of PRKAG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRKAG1 | 5571 | 6 | 1 | 23 | |
| GSE20123 | PRKAG1 | 5571 | 6 | 1 | 23 | |
| GSE43470 | PRKAG1 | 5571 | 4 | 1 | 38 | |
| GSE46452 | PRKAG1 | 5571 | 8 | 1 | 50 | |
| GSE47630 | PRKAG1 | 5571 | 11 | 2 | 27 | |
| GSE54993 | PRKAG1 | 5571 | 0 | 5 | 65 | |
| GSE54994 | PRKAG1 | 5571 | 4 | 1 | 48 | |
| GSE60625 | PRKAG1 | 5571 | 0 | 0 | 11 | |
| GSE74703 | PRKAG1 | 5571 | 4 | 1 | 31 | |
| GSE74704 | PRKAG1 | 5571 | 4 | 1 | 15 | |
| TCGA | PRKAG1 | 5571 | 16 | 15 | 65 |
Total number of gains: 63; Total number of losses: 29; Total Number of normals: 396.
Somatic mutations of PRKAG1:
Generating mutation plots.
Highly correlated genes for PRKAG1:
Showing top 20/653 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRKAG1 | DLG3 | 0.797082 | 3 | 0 | 3 |
| PRKAG1 | SLTM | 0.783474 | 3 | 0 | 3 |
| PRKAG1 | DHX35 | 0.778084 | 3 | 0 | 3 |
| PRKAG1 | TOPORS | 0.776191 | 3 | 0 | 3 |
| PRKAG1 | KIAA0556 | 0.764923 | 4 | 0 | 4 |
| PRKAG1 | NIPSNAP3A | 0.761864 | 4 | 0 | 4 |
| PRKAG1 | TMEM218 | 0.761351 | 4 | 0 | 4 |
| PRKAG1 | CGNL1 | 0.75799 | 3 | 0 | 3 |
| PRKAG1 | ESYT2 | 0.753789 | 3 | 0 | 3 |
| PRKAG1 | INPP1 | 0.753572 | 3 | 0 | 3 |
| PRKAG1 | TMEM80 | 0.750732 | 3 | 0 | 3 |
| PRKAG1 | NUB1 | 0.748499 | 3 | 0 | 3 |
| PRKAG1 | THAP2 | 0.735254 | 3 | 0 | 3 |
| PRKAG1 | SLC35C2 | 0.733221 | 3 | 0 | 3 |
| PRKAG1 | TMEM205 | 0.731935 | 3 | 0 | 3 |
| PRKAG1 | FEM1A | 0.730948 | 3 | 0 | 3 |
| PRKAG1 | PRPF38A | 0.72788 | 5 | 0 | 5 |
| PRKAG1 | TMEM50B | 0.724468 | 5 | 0 | 5 |
| PRKAG1 | ALKBH8 | 0.724113 | 3 | 0 | 3 |
| PRKAG1 | FGFR2 | 0.721962 | 3 | 0 | 3 |
For details and further investigation, click here