| Full name: tripartite motif containing 33 | Alias Symbol: TIF1GAMMA|FLJ11429|KIAA1113|TIFGAMMA|RFG7|TF1G|TIF1G|PTC7 | ||
| Type: protein-coding gene | Cytoband: 1p13.2 | ||
| Entrez ID: 51592 | HGNC ID: HGNC:16290 | Ensembl Gene: ENSG00000197323 | OMIM ID: 605769 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRIM33:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM33 | 51592 | 210266_s_at | -0.2246 | 0.5062 | |
| GSE20347 | TRIM33 | 51592 | 210266_s_at | -0.2922 | 0.1473 | |
| GSE23400 | TRIM33 | 51592 | 210266_s_at | -0.2049 | 0.0014 | |
| GSE26886 | TRIM33 | 51592 | 212435_at | -0.6730 | 0.0018 | |
| GSE29001 | TRIM33 | 51592 | 210266_s_at | -0.3718 | 0.2146 | |
| GSE38129 | TRIM33 | 51592 | 210266_s_at | -0.2367 | 0.1415 | |
| GSE45670 | TRIM33 | 51592 | 210266_s_at | -0.1703 | 0.2821 | |
| GSE53622 | TRIM33 | 51592 | 8797 | -0.6248 | 0.0000 | |
| GSE53624 | TRIM33 | 51592 | 8797 | -0.4355 | 0.0000 | |
| GSE63941 | TRIM33 | 51592 | 212435_at | 0.2821 | 0.6974 | |
| GSE77861 | TRIM33 | 51592 | 210266_s_at | -0.0701 | 0.8101 | |
| GSE97050 | TRIM33 | 51592 | A_33_P3257367 | -0.5421 | 0.1095 | |
| SRP007169 | TRIM33 | 51592 | RNAseq | -0.0330 | 0.9290 | |
| SRP008496 | TRIM33 | 51592 | RNAseq | -0.0994 | 0.5878 | |
| SRP064894 | TRIM33 | 51592 | RNAseq | -0.3013 | 0.1050 | |
| SRP133303 | TRIM33 | 51592 | RNAseq | -0.1378 | 0.1550 | |
| SRP159526 | TRIM33 | 51592 | RNAseq | -0.7035 | 0.0067 | |
| SRP193095 | TRIM33 | 51592 | RNAseq | -0.3353 | 0.0000 | |
| SRP219564 | TRIM33 | 51592 | RNAseq | -0.6703 | 0.1289 | |
| TCGA | TRIM33 | 51592 | RNAseq | -0.0729 | 0.1122 |
Upregulated datasets: 0; Downregulated datasets: 0.
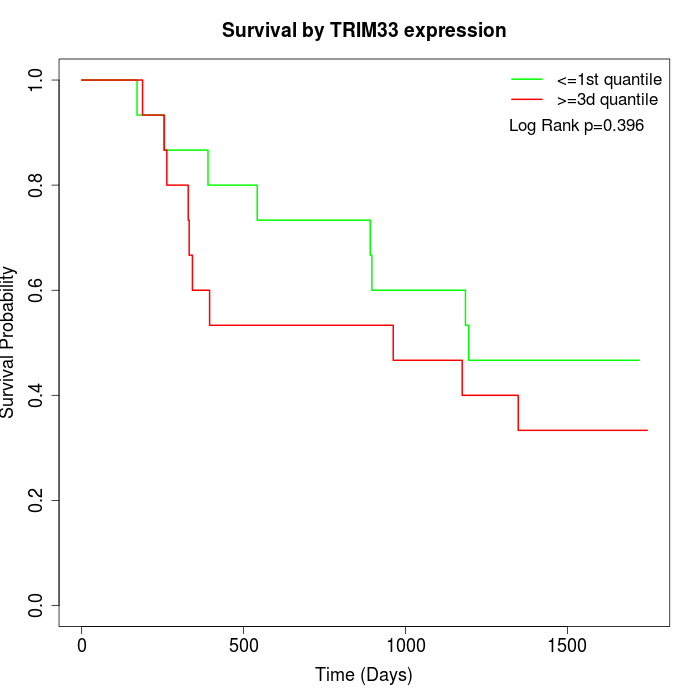
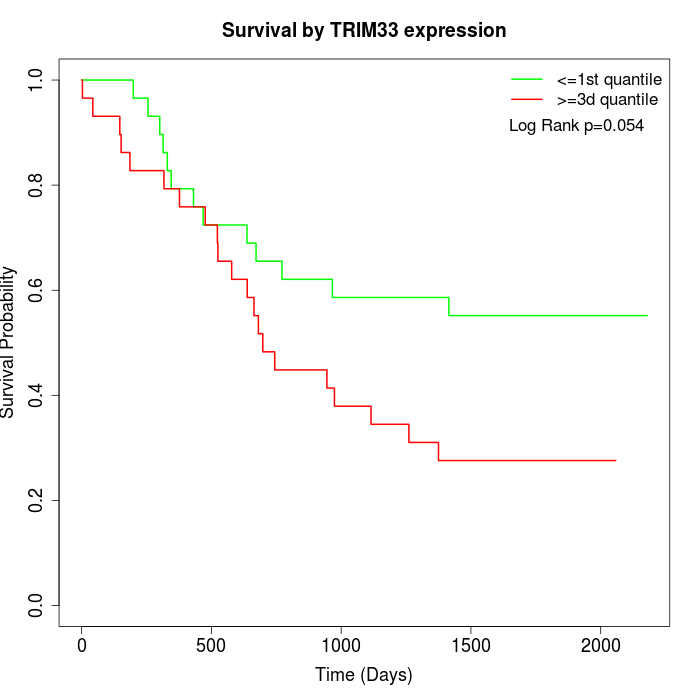
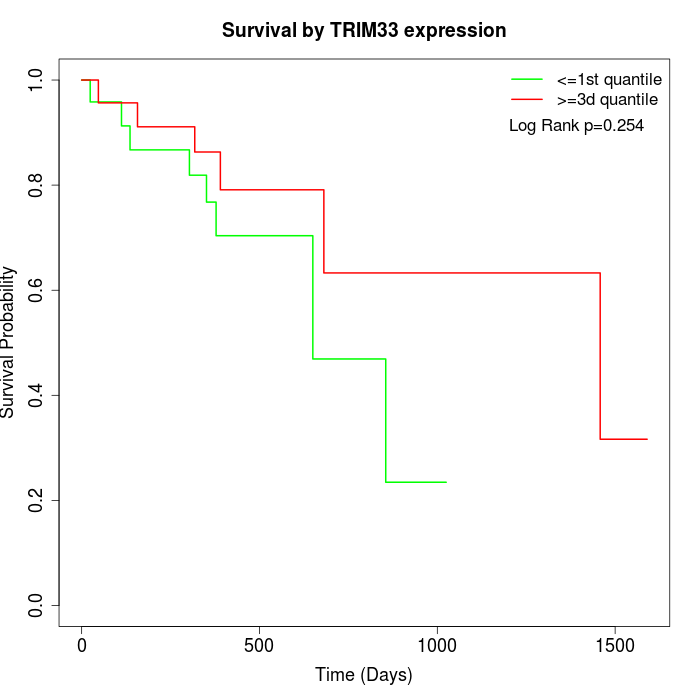
Survival by TRIM33 expression:
Note: Click image to view full size file.
Copy number change of TRIM33:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM33 | 51592 | 0 | 9 | 21 | |
| GSE20123 | TRIM33 | 51592 | 0 | 9 | 21 | |
| GSE43470 | TRIM33 | 51592 | 0 | 8 | 35 | |
| GSE46452 | TRIM33 | 51592 | 2 | 1 | 56 | |
| GSE47630 | TRIM33 | 51592 | 9 | 5 | 26 | |
| GSE54993 | TRIM33 | 51592 | 0 | 1 | 69 | |
| GSE54994 | TRIM33 | 51592 | 7 | 3 | 43 | |
| GSE60625 | TRIM33 | 51592 | 0 | 0 | 11 | |
| GSE74703 | TRIM33 | 51592 | 0 | 7 | 29 | |
| GSE74704 | TRIM33 | 51592 | 0 | 5 | 15 | |
| TCGA | TRIM33 | 51592 | 11 | 30 | 55 |
Total number of gains: 29; Total number of losses: 78; Total Number of normals: 381.
Somatic mutations of TRIM33:
Generating mutation plots.
Highly correlated genes for TRIM33:
Showing top 20/676 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM33 | RNF207 | 0.764646 | 4 | 0 | 4 |
| TRIM33 | CRBN | 0.734932 | 3 | 0 | 3 |
| TRIM33 | IGIP | 0.731262 | 4 | 0 | 4 |
| TRIM33 | FAM53B | 0.724678 | 3 | 0 | 3 |
| TRIM33 | MOCS2 | 0.724213 | 3 | 0 | 3 |
| TRIM33 | SSNA1 | 0.723662 | 3 | 0 | 3 |
| TRIM33 | ZACN | 0.721878 | 3 | 0 | 3 |
| TRIM33 | ALKBH7 | 0.715896 | 3 | 0 | 3 |
| TRIM33 | HDHD2 | 0.715312 | 3 | 0 | 3 |
| TRIM33 | RWDD2B | 0.713594 | 4 | 0 | 3 |
| TRIM33 | AKAP17A | 0.703532 | 3 | 0 | 3 |
| TRIM33 | RPS14 | 0.703443 | 3 | 0 | 3 |
| TRIM33 | ZNF571 | 0.701928 | 4 | 0 | 3 |
| TRIM33 | FAM122A | 0.700706 | 4 | 0 | 4 |
| TRIM33 | SNX4 | 0.700588 | 4 | 0 | 3 |
| TRIM33 | ANKRA2 | 0.699651 | 3 | 0 | 3 |
| TRIM33 | CCDC7 | 0.699317 | 3 | 0 | 3 |
| TRIM33 | ATG14 | 0.698229 | 3 | 0 | 3 |
| TRIM33 | DNAJB12 | 0.696714 | 4 | 0 | 4 |
| TRIM33 | FAM174A | 0.690497 | 4 | 0 | 3 |
For details and further investigation, click here