| Full name: dual specificity phosphatase 10 | Alias Symbol: MKP-5|MKP5 | ||
| Type: protein-coding gene | Cytoband: 1q41 | ||
| Entrez ID: 11221 | HGNC ID: HGNC:3065 | Ensembl Gene: ENSG00000143507 | OMIM ID: 608867 |
| Drug and gene relationship at DGIdb | |||
DUSP10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway |
Expression of DUSP10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DUSP10 | 11221 | 221563_at | 0.0695 | 0.9466 | |
| GSE20347 | DUSP10 | 11221 | 221563_at | 0.9536 | 0.0019 | |
| GSE23400 | DUSP10 | 11221 | 221563_at | 0.3108 | 0.0012 | |
| GSE26886 | DUSP10 | 11221 | 221563_at | -0.1112 | 0.7448 | |
| GSE29001 | DUSP10 | 11221 | 221563_at | 0.3782 | 0.4126 | |
| GSE38129 | DUSP10 | 11221 | 221563_at | 0.8131 | 0.0004 | |
| GSE45670 | DUSP10 | 11221 | 221563_at | 0.9148 | 0.0020 | |
| GSE53622 | DUSP10 | 11221 | 48432 | 0.8512 | 0.0000 | |
| GSE53624 | DUSP10 | 11221 | 48432 | 0.9063 | 0.0000 | |
| GSE63941 | DUSP10 | 11221 | 221563_at | -2.1457 | 0.0624 | |
| GSE77861 | DUSP10 | 11221 | 221563_at | 1.0771 | 0.0089 | |
| GSE97050 | DUSP10 | 11221 | A_24_P182494 | -0.2193 | 0.7743 | |
| SRP007169 | DUSP10 | 11221 | RNAseq | 0.6572 | 0.2117 | |
| SRP008496 | DUSP10 | 11221 | RNAseq | 1.2299 | 0.0081 | |
| SRP064894 | DUSP10 | 11221 | RNAseq | 1.3241 | 0.0000 | |
| SRP133303 | DUSP10 | 11221 | RNAseq | 1.3151 | 0.0003 | |
| SRP159526 | DUSP10 | 11221 | RNAseq | 0.1937 | 0.5916 | |
| SRP193095 | DUSP10 | 11221 | RNAseq | 1.6501 | 0.0000 | |
| SRP219564 | DUSP10 | 11221 | RNAseq | 0.6356 | 0.3280 | |
| TCGA | DUSP10 | 11221 | RNAseq | 0.4748 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
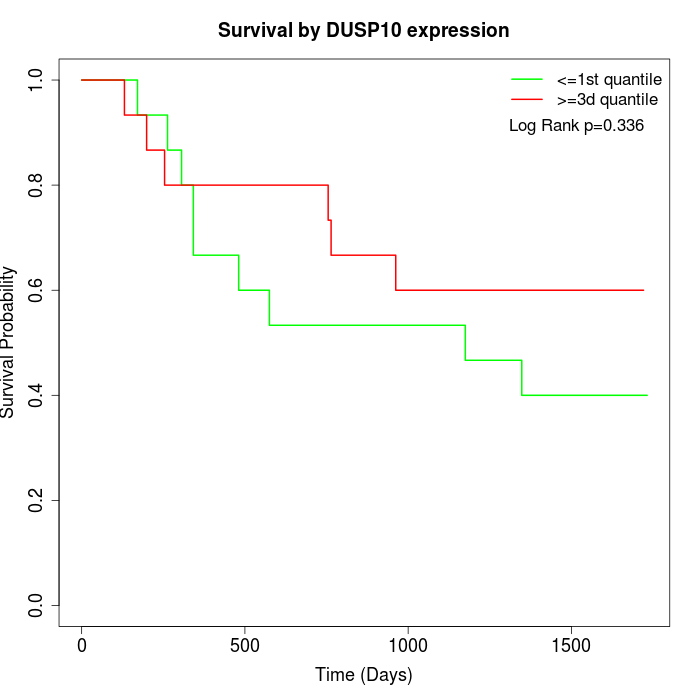
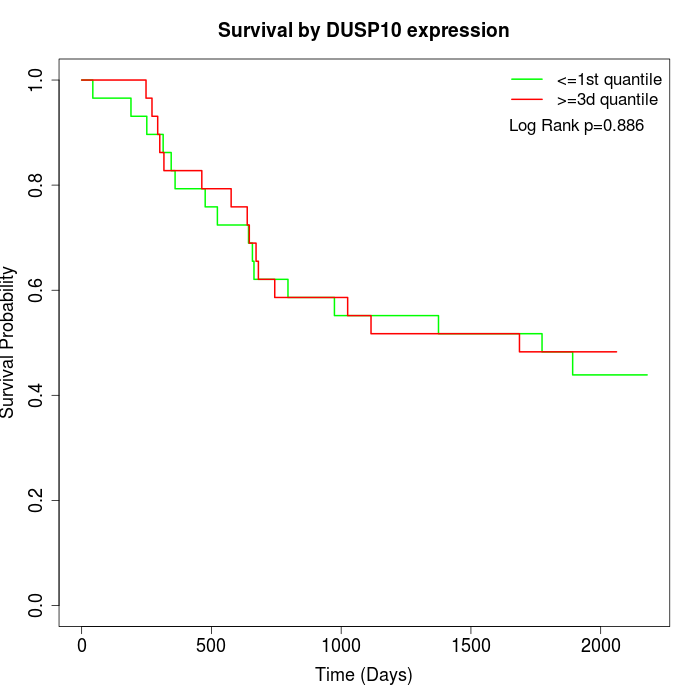
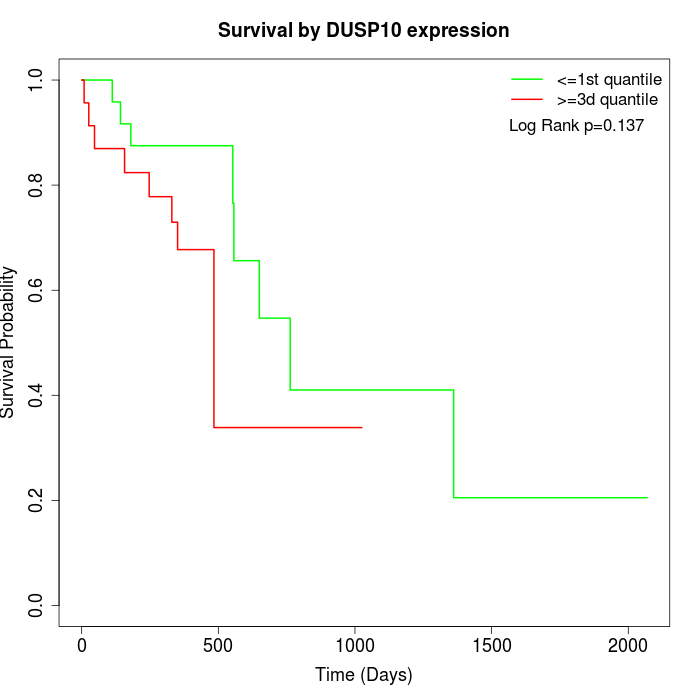
Survival by DUSP10 expression:
Note: Click image to view full size file.
Copy number change of DUSP10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DUSP10 | 11221 | 10 | 0 | 20 | |
| GSE20123 | DUSP10 | 11221 | 10 | 0 | 20 | |
| GSE43470 | DUSP10 | 11221 | 7 | 0 | 36 | |
| GSE46452 | DUSP10 | 11221 | 3 | 1 | 55 | |
| GSE47630 | DUSP10 | 11221 | 15 | 0 | 25 | |
| GSE54993 | DUSP10 | 11221 | 0 | 6 | 64 | |
| GSE54994 | DUSP10 | 11221 | 16 | 0 | 37 | |
| GSE60625 | DUSP10 | 11221 | 0 | 0 | 11 | |
| GSE74703 | DUSP10 | 11221 | 7 | 0 | 29 | |
| GSE74704 | DUSP10 | 11221 | 4 | 0 | 16 | |
| TCGA | DUSP10 | 11221 | 45 | 3 | 48 |
Total number of gains: 117; Total number of losses: 10; Total Number of normals: 361.
Somatic mutations of DUSP10:
Generating mutation plots.
Highly correlated genes for DUSP10:
Showing top 20/292 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DUSP10 | IMPAD1 | 0.665505 | 3 | 0 | 3 |
| DUSP10 | AZIN2 | 0.646462 | 3 | 0 | 3 |
| DUSP10 | TTYH3 | 0.645533 | 4 | 0 | 4 |
| DUSP10 | WDR66 | 0.643061 | 4 | 0 | 3 |
| DUSP10 | FBLIM1 | 0.641438 | 4 | 0 | 3 |
| DUSP10 | RBP1 | 0.634956 | 7 | 0 | 7 |
| DUSP10 | B3GNT9 | 0.634699 | 5 | 0 | 5 |
| DUSP10 | SLC20A2 | 0.630602 | 3 | 0 | 3 |
| DUSP10 | SERPINH1 | 0.621038 | 7 | 0 | 6 |
| DUSP10 | GALNT18 | 0.620206 | 4 | 0 | 4 |
| DUSP10 | TMEM155 | 0.619221 | 3 | 0 | 3 |
| DUSP10 | GDI1 | 0.615527 | 4 | 0 | 3 |
| DUSP10 | SH3PXD2B | 0.614721 | 5 | 0 | 4 |
| DUSP10 | SHC4 | 0.613413 | 4 | 0 | 3 |
| DUSP10 | MYO5A | 0.610712 | 7 | 0 | 6 |
| DUSP10 | YTHDF1 | 0.61008 | 3 | 0 | 3 |
| DUSP10 | RUNX2 | 0.607353 | 3 | 0 | 3 |
| DUSP10 | WDR54 | 0.607258 | 4 | 0 | 4 |
| DUSP10 | MEF2A | 0.607107 | 5 | 0 | 4 |
| DUSP10 | FMNL2 | 0.606708 | 5 | 0 | 5 |
For details and further investigation, click here