| Full name: ETS transcription factor ELK1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp11.23 | ||
| Entrez ID: 2002 | HGNC ID: HGNC:3321 | Ensembl Gene: ENSG00000126767 | OMIM ID: 311040 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ELK1 involved pathways:
Expression of ELK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ELK1 | 2002 | 203617_x_at | 0.3581 | 0.2526 | |
| GSE20347 | ELK1 | 2002 | 203617_x_at | 0.1717 | 0.0198 | |
| GSE23400 | ELK1 | 2002 | 203617_x_at | 0.0699 | 0.0853 | |
| GSE26886 | ELK1 | 2002 | 203617_x_at | 0.3758 | 0.0048 | |
| GSE29001 | ELK1 | 2002 | 203617_x_at | -0.1911 | 0.1684 | |
| GSE38129 | ELK1 | 2002 | 203617_x_at | 0.1704 | 0.0798 | |
| GSE45670 | ELK1 | 2002 | 203617_x_at | 0.1938 | 0.2182 | |
| GSE53622 | ELK1 | 2002 | 3827 | 0.3744 | 0.0000 | |
| GSE53624 | ELK1 | 2002 | 123917 | 0.7695 | 0.0000 | |
| GSE63941 | ELK1 | 2002 | 203617_x_at | -0.2423 | 0.3776 | |
| GSE77861 | ELK1 | 2002 | 203617_x_at | 0.1114 | 0.4440 | |
| GSE97050 | ELK1 | 2002 | A_33_P3386099 | 0.1514 | 0.4700 | |
| SRP007169 | ELK1 | 2002 | RNAseq | 0.6009 | 0.1269 | |
| SRP008496 | ELK1 | 2002 | RNAseq | 0.6327 | 0.0095 | |
| SRP064894 | ELK1 | 2002 | RNAseq | 0.5424 | 0.0030 | |
| SRP133303 | ELK1 | 2002 | RNAseq | 0.6676 | 0.0057 | |
| SRP159526 | ELK1 | 2002 | RNAseq | 0.8368 | 0.0000 | |
| SRP193095 | ELK1 | 2002 | RNAseq | 0.5660 | 0.0000 | |
| SRP219564 | ELK1 | 2002 | RNAseq | 0.4433 | 0.2201 | |
| TCGA | ELK1 | 2002 | RNAseq | 0.1600 | 0.0030 |
Upregulated datasets: 0; Downregulated datasets: 0.
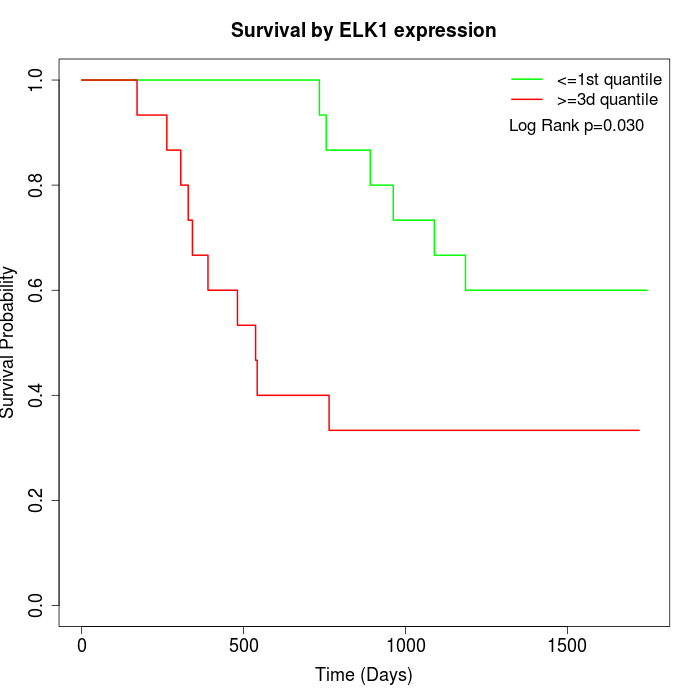
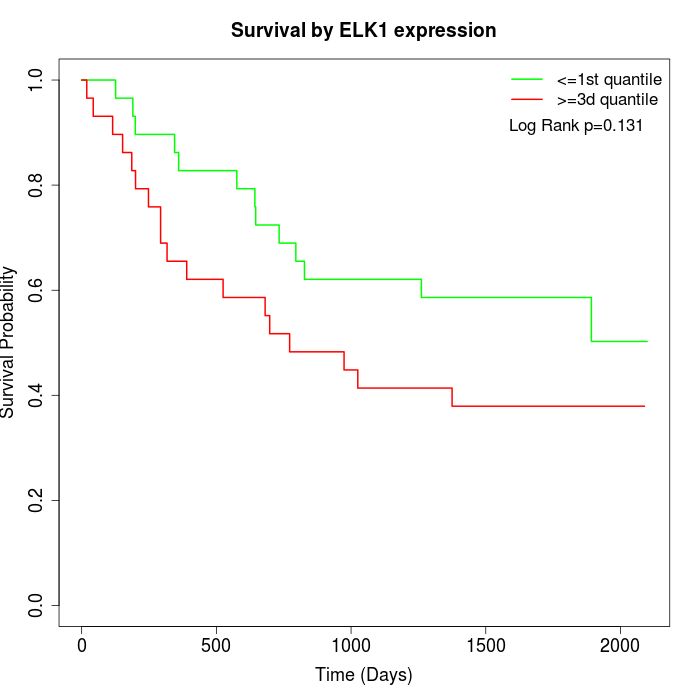
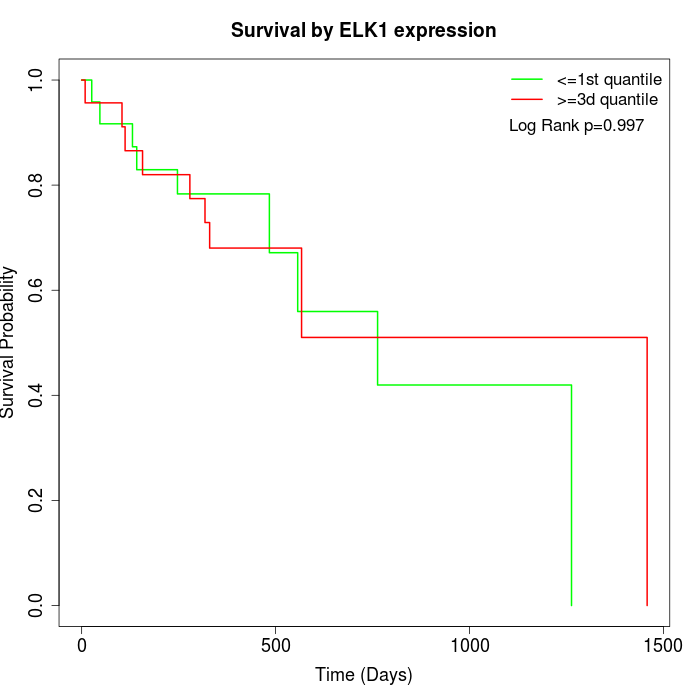
Survival by ELK1 expression:
Note: Click image to view full size file.
Copy number change of ELK1:
No record found for this gene.
Somatic mutations of ELK1:
Generating mutation plots.
Highly correlated genes for ELK1:
Showing top 20/299 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ELK1 | GJA4 | 0.77197 | 3 | 0 | 3 |
| ELK1 | SLC45A3 | 0.730889 | 3 | 0 | 3 |
| ELK1 | STK11 | 0.713984 | 3 | 0 | 3 |
| ELK1 | DMTN | 0.693555 | 3 | 0 | 3 |
| ELK1 | C9orf16 | 0.693219 | 4 | 0 | 4 |
| ELK1 | CTF1 | 0.682712 | 3 | 0 | 3 |
| ELK1 | WDR24 | 0.673342 | 4 | 0 | 3 |
| ELK1 | TBC1D25 | 0.65569 | 4 | 0 | 3 |
| ELK1 | YDJC | 0.6516 | 5 | 0 | 5 |
| ELK1 | PPP2R5B | 0.648349 | 7 | 0 | 6 |
| ELK1 | AP2A1 | 0.644897 | 3 | 0 | 3 |
| ELK1 | TAOK2 | 0.642611 | 4 | 0 | 3 |
| ELK1 | BMP8A | 0.639477 | 4 | 0 | 3 |
| ELK1 | KDM6B | 0.636699 | 3 | 0 | 3 |
| ELK1 | ANKRD52 | 0.635182 | 7 | 0 | 5 |
| ELK1 | CAPN15 | 0.634704 | 3 | 0 | 3 |
| ELK1 | MCM3AP | 0.628725 | 3 | 0 | 3 |
| ELK1 | FMNL1 | 0.627392 | 4 | 0 | 4 |
| ELK1 | CHD5 | 0.62684 | 3 | 0 | 3 |
| ELK1 | RHEBL1 | 0.625452 | 4 | 0 | 3 |
For details and further investigation, click here