| Full name: enolase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.31 | ||
| Entrez ID: 2026 | HGNC ID: HGNC:3353 | Ensembl Gene: ENSG00000111674 | OMIM ID: 131360 |
| Drug and gene relationship at DGIdb | |||
ENO2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of ENO2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ENO2 | 2026 | 201313_at | 0.1508 | 0.9135 | |
| GSE20347 | ENO2 | 2026 | 201313_at | 1.8692 | 0.0000 | |
| GSE23400 | ENO2 | 2026 | 201313_at | 0.9736 | 0.0000 | |
| GSE26886 | ENO2 | 2026 | 201313_at | 2.2760 | 0.0000 | |
| GSE29001 | ENO2 | 2026 | 201313_at | 1.4947 | 0.0035 | |
| GSE38129 | ENO2 | 2026 | 201313_at | 1.2697 | 0.0007 | |
| GSE45670 | ENO2 | 2026 | 201313_at | 0.9800 | 0.0911 | |
| GSE53622 | ENO2 | 2026 | 115072 | 1.2158 | 0.0000 | |
| GSE53624 | ENO2 | 2026 | 115072 | 1.4963 | 0.0000 | |
| GSE63941 | ENO2 | 2026 | 201313_at | 0.3430 | 0.7605 | |
| GSE77861 | ENO2 | 2026 | 201313_at | 1.9557 | 0.0329 | |
| GSE97050 | ENO2 | 2026 | A_24_P236091 | -0.5021 | 0.4176 | |
| SRP007169 | ENO2 | 2026 | RNAseq | 4.0334 | 0.0000 | |
| SRP008496 | ENO2 | 2026 | RNAseq | 3.3033 | 0.0000 | |
| SRP064894 | ENO2 | 2026 | RNAseq | 1.4830 | 0.0000 | |
| SRP133303 | ENO2 | 2026 | RNAseq | 1.8586 | 0.0000 | |
| SRP159526 | ENO2 | 2026 | RNAseq | 1.3256 | 0.0001 | |
| SRP193095 | ENO2 | 2026 | RNAseq | 2.5667 | 0.0000 | |
| SRP219564 | ENO2 | 2026 | RNAseq | 0.3020 | 0.5818 | |
| TCGA | ENO2 | 2026 | RNAseq | 0.2687 | 0.0413 |
Upregulated datasets: 13; Downregulated datasets: 0.
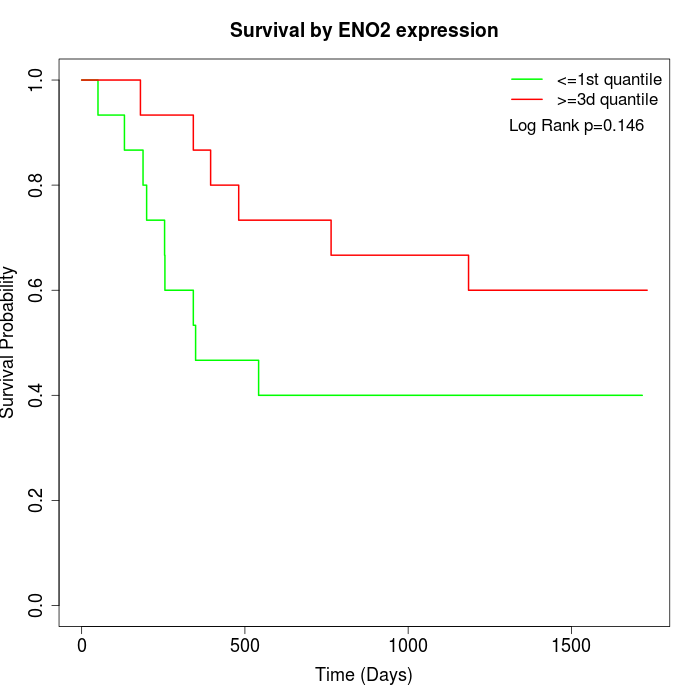
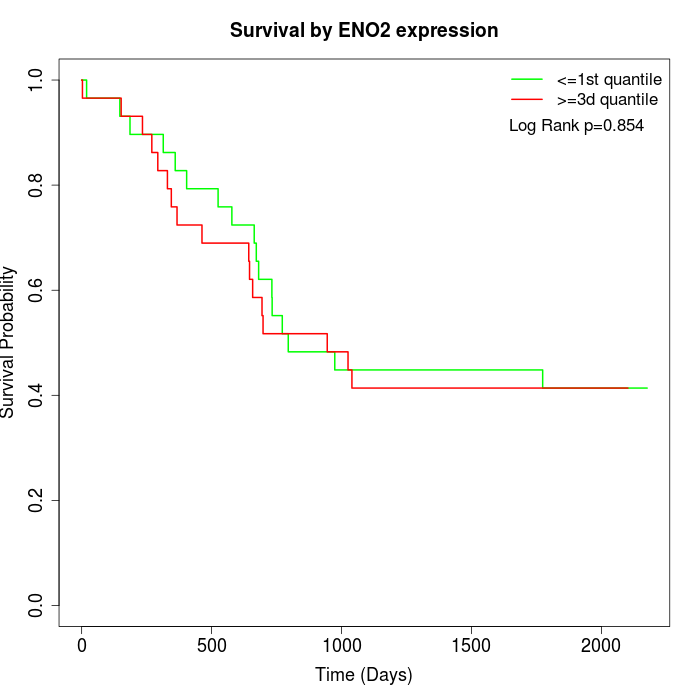
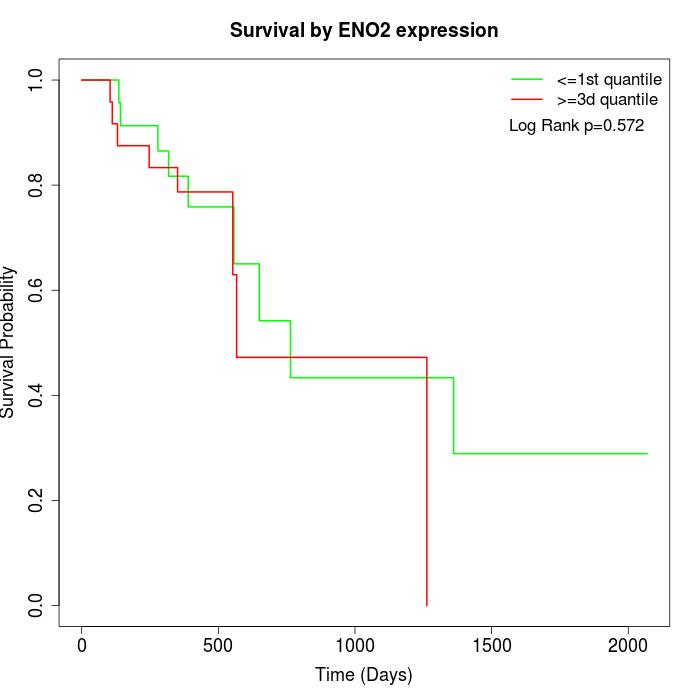
Survival by ENO2 expression:
Note: Click image to view full size file.
Copy number change of ENO2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ENO2 | 2026 | 9 | 3 | 18 | |
| GSE20123 | ENO2 | 2026 | 9 | 3 | 18 | |
| GSE43470 | ENO2 | 2026 | 8 | 3 | 32 | |
| GSE46452 | ENO2 | 2026 | 10 | 1 | 48 | |
| GSE47630 | ENO2 | 2026 | 12 | 2 | 26 | |
| GSE54993 | ENO2 | 2026 | 1 | 10 | 59 | |
| GSE54994 | ENO2 | 2026 | 10 | 2 | 41 | |
| GSE60625 | ENO2 | 2026 | 0 | 1 | 10 | |
| GSE74703 | ENO2 | 2026 | 7 | 2 | 27 | |
| GSE74704 | ENO2 | 2026 | 5 | 3 | 12 | |
| TCGA | ENO2 | 2026 | 40 | 6 | 50 |
Total number of gains: 111; Total number of losses: 36; Total Number of normals: 341.
Somatic mutations of ENO2:
Generating mutation plots.
Highly correlated genes for ENO2:
Showing top 20/1583 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ENO2 | DCHS2 | 0.795627 | 3 | 0 | 3 |
| ENO2 | ZBED1 | 0.793367 | 3 | 0 | 3 |
| ENO2 | TCTN1 | 0.783056 | 3 | 0 | 3 |
| ENO2 | AURKAIP1 | 0.779422 | 3 | 0 | 3 |
| ENO2 | FADS1 | 0.77105 | 3 | 0 | 3 |
| ENO2 | SSBP4 | 0.763488 | 4 | 0 | 4 |
| ENO2 | GLIPR2 | 0.754622 | 3 | 0 | 3 |
| ENO2 | ALPK3 | 0.750926 | 3 | 0 | 3 |
| ENO2 | NT5M | 0.747585 | 4 | 0 | 4 |
| ENO2 | FILIP1 | 0.742295 | 4 | 0 | 4 |
| ENO2 | S1PR3 | 0.734847 | 4 | 0 | 4 |
| ENO2 | C11orf96 | 0.732877 | 3 | 0 | 3 |
| ENO2 | LIX1L | 0.731238 | 4 | 0 | 4 |
| ENO2 | HMCN1 | 0.730198 | 4 | 0 | 3 |
| ENO2 | KRTCAP2 | 0.729711 | 4 | 0 | 4 |
| ENO2 | FNIP2 | 0.728043 | 3 | 0 | 3 |
| ENO2 | DOCK11 | 0.724124 | 4 | 0 | 4 |
| ENO2 | PKIG | 0.72397 | 5 | 0 | 5 |
| ENO2 | RILPL1 | 0.718118 | 4 | 0 | 4 |
| ENO2 | CRTC2 | 0.717253 | 4 | 0 | 4 |
For details and further investigation, click here