| Full name: fetal and adult testis expressed 1 | Alias Symbol: FATE|CT43 | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 89885 | HGNC ID: HGNC:24683 | Ensembl Gene: ENSG00000147378 | OMIM ID: 300450 |
| Drug and gene relationship at DGIdb | |||
Expression of FATE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FATE1 | 89885 | 231573_at | -0.1704 | 0.5078 | |
| GSE26886 | FATE1 | 89885 | 231573_at | -0.0193 | 0.8841 | |
| GSE45670 | FATE1 | 89885 | 231573_at | 0.1192 | 0.4197 | |
| GSE53622 | FATE1 | 89885 | 27433 | 0.2939 | 0.0000 | |
| GSE53624 | FATE1 | 89885 | 27433 | 0.3863 | 0.0000 | |
| GSE63941 | FATE1 | 89885 | 231573_at | -0.0464 | 0.8162 | |
| GSE77861 | FATE1 | 89885 | 231573_at | -0.2766 | 0.0293 | |
| TCGA | FATE1 | 89885 | RNAseq | 0.5413 | 0.4831 |
Upregulated datasets: 0; Downregulated datasets: 0.
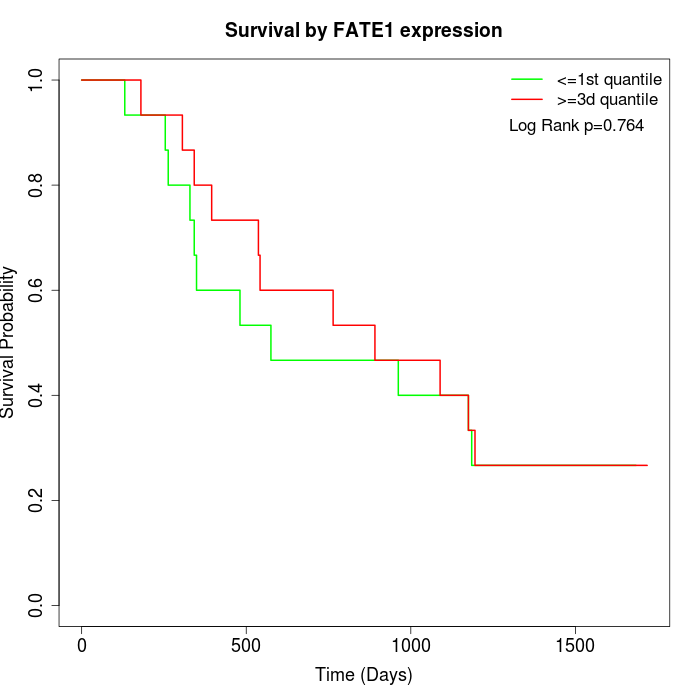
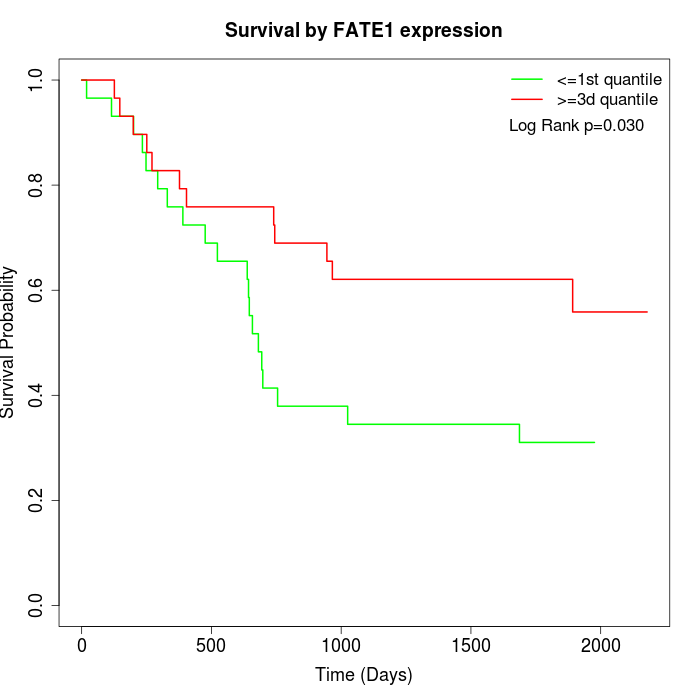
Survival by FATE1 expression:
Note: Click image to view full size file.
Copy number change of FATE1:
No record found for this gene.
Somatic mutations of FATE1:
Generating mutation plots.
Highly correlated genes for FATE1:
Showing top 20/226 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FATE1 | NAALADL1 | 0.738586 | 3 | 0 | 3 |
| FATE1 | C4BPB | 0.71748 | 3 | 0 | 3 |
| FATE1 | PSPN | 0.690676 | 3 | 0 | 3 |
| FATE1 | ZFHX2 | 0.672118 | 5 | 0 | 5 |
| FATE1 | MVK | 0.671569 | 3 | 0 | 3 |
| FATE1 | TSGA10IP | 0.66946 | 4 | 0 | 4 |
| FATE1 | TNR | 0.666355 | 3 | 0 | 3 |
| FATE1 | NHLRC4 | 0.665697 | 4 | 0 | 4 |
| FATE1 | CYP2A7 | 0.664126 | 3 | 0 | 3 |
| FATE1 | CCDC13 | 0.663266 | 3 | 0 | 3 |
| FATE1 | ZBTB20-AS1 | 0.663259 | 3 | 0 | 3 |
| FATE1 | SPPL2C | 0.660392 | 3 | 0 | 3 |
| FATE1 | BPIFA2 | 0.65859 | 3 | 0 | 3 |
| FATE1 | RASSF7 | 0.649709 | 3 | 0 | 3 |
| FATE1 | KIAA1614 | 0.648259 | 3 | 0 | 3 |
| FATE1 | CST8 | 0.646225 | 3 | 0 | 3 |
| FATE1 | TSSK2 | 0.645599 | 4 | 0 | 3 |
| FATE1 | VGLL2 | 0.644425 | 4 | 0 | 3 |
| FATE1 | CNR2 | 0.641816 | 4 | 0 | 3 |
| FATE1 | B3GAT1 | 0.638086 | 3 | 0 | 3 |
For details and further investigation, click here