| Full name: beta-1,3-glucuronyltransferase 1 | Alias Symbol: GlcAT-P|HNK-1|NK-1 | ||
| Type: protein-coding gene | Cytoband: 11q25 | ||
| Entrez ID: 27087 | HGNC ID: HGNC:921 | Ensembl Gene: ENSG00000109956 | OMIM ID: 151290 |
| Drug and gene relationship at DGIdb | |||
Expression of B3GAT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | B3GAT1 | 27087 | 219521_at | -0.0318 | 0.9135 | |
| GSE20347 | B3GAT1 | 27087 | 219521_at | 0.0113 | 0.9178 | |
| GSE23400 | B3GAT1 | 27087 | 219521_at | -0.1512 | 0.0028 | |
| GSE26886 | B3GAT1 | 27087 | 219521_at | 0.0829 | 0.5034 | |
| GSE29001 | B3GAT1 | 27087 | 219521_at | -0.0040 | 0.9835 | |
| GSE38129 | B3GAT1 | 27087 | 219521_at | -0.1791 | 0.1333 | |
| GSE45670 | B3GAT1 | 27087 | 219521_at | 0.1698 | 0.0629 | |
| GSE53622 | B3GAT1 | 27087 | 96669 | -0.1375 | 0.5309 | |
| GSE53624 | B3GAT1 | 27087 | 96669 | -0.2293 | 0.1869 | |
| GSE63941 | B3GAT1 | 27087 | 219521_at | 0.0861 | 0.7789 | |
| GSE77861 | B3GAT1 | 27087 | 219521_at | -0.0691 | 0.5848 | |
| SRP219564 | B3GAT1 | 27087 | RNAseq | 1.0548 | 0.0971 | |
| TCGA | B3GAT1 | 27087 | RNAseq | -3.1980 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
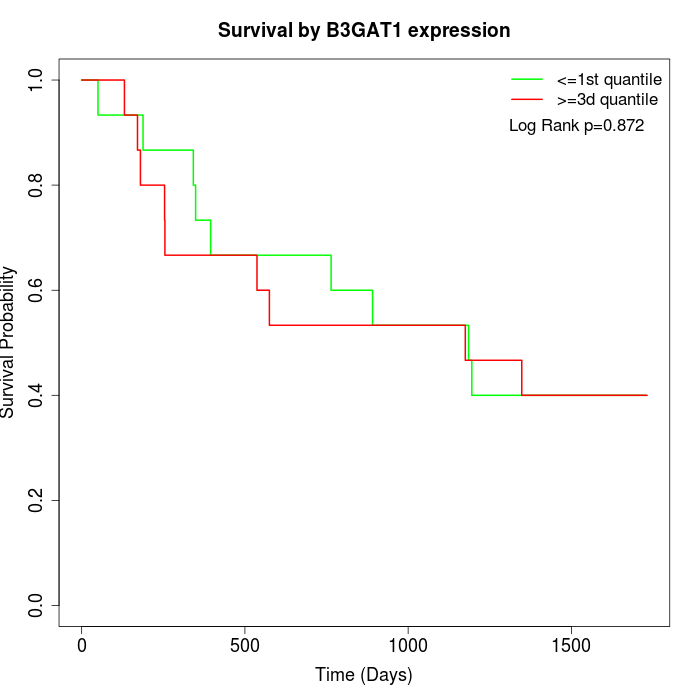
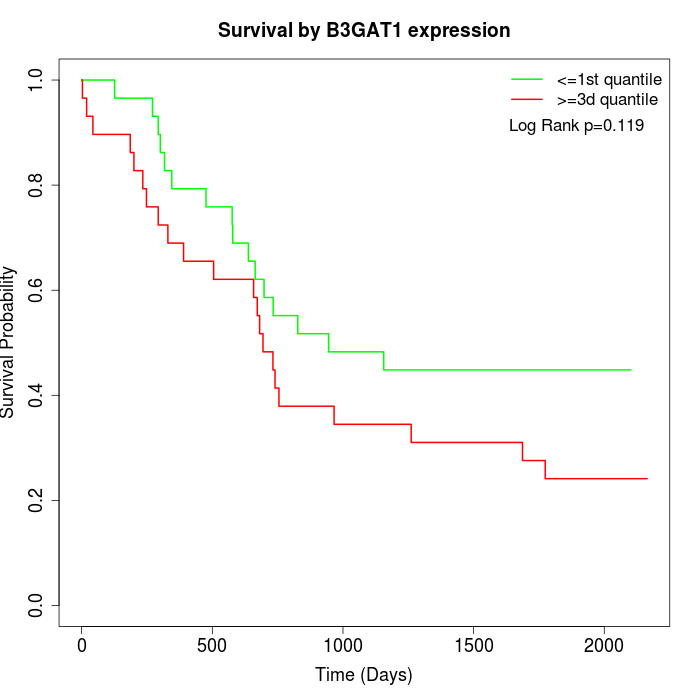
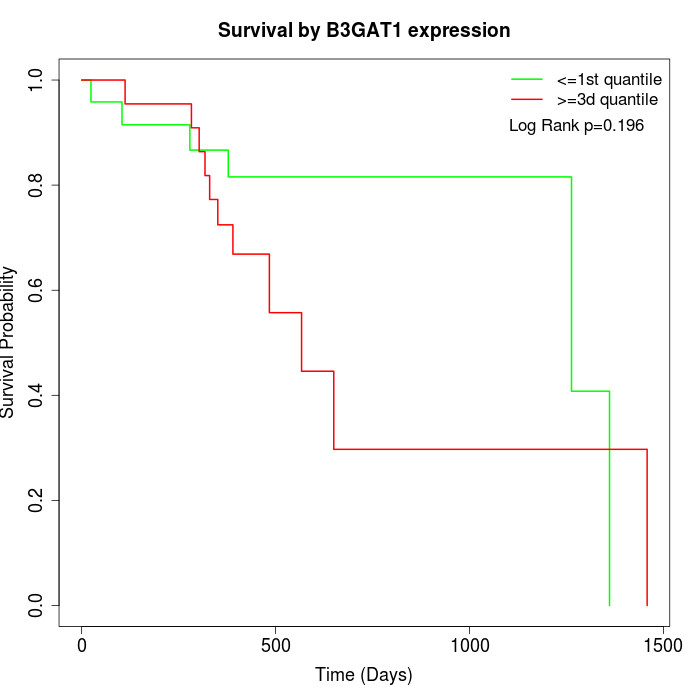
Survival by B3GAT1 expression:
Note: Click image to view full size file.
Copy number change of B3GAT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | B3GAT1 | 27087 | 0 | 15 | 15 | |
| GSE20123 | B3GAT1 | 27087 | 0 | 15 | 15 | |
| GSE43470 | B3GAT1 | 27087 | 2 | 8 | 33 | |
| GSE46452 | B3GAT1 | 27087 | 3 | 26 | 30 | |
| GSE47630 | B3GAT1 | 27087 | 3 | 18 | 19 | |
| GSE54993 | B3GAT1 | 27087 | 10 | 0 | 60 | |
| GSE54994 | B3GAT1 | 27087 | 5 | 18 | 30 | |
| GSE60625 | B3GAT1 | 27087 | 0 | 3 | 8 | |
| GSE74703 | B3GAT1 | 27087 | 1 | 5 | 30 | |
| GSE74704 | B3GAT1 | 27087 | 0 | 11 | 9 | |
| TCGA | B3GAT1 | 27087 | 6 | 49 | 41 |
Total number of gains: 30; Total number of losses: 168; Total Number of normals: 290.
Somatic mutations of B3GAT1:
Generating mutation plots.
Highly correlated genes for B3GAT1:
Showing top 20/728 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| B3GAT1 | ATG10 | 0.678348 | 4 | 0 | 4 |
| B3GAT1 | RECK | 0.667789 | 3 | 0 | 3 |
| B3GAT1 | IL17A | 0.663266 | 5 | 0 | 4 |
| B3GAT1 | GCG | 0.658176 | 3 | 0 | 3 |
| B3GAT1 | CDC25C | 0.656206 | 3 | 0 | 3 |
| B3GAT1 | NCR2 | 0.655951 | 6 | 0 | 5 |
| B3GAT1 | KCNE2 | 0.649304 | 6 | 0 | 6 |
| B3GAT1 | GPLD1 | 0.648764 | 3 | 0 | 3 |
| B3GAT1 | GRK6 | 0.648026 | 5 | 0 | 5 |
| B3GAT1 | MUC5AC | 0.647862 | 4 | 0 | 4 |
| B3GAT1 | RNF17 | 0.647048 | 4 | 0 | 4 |
| B3GAT1 | LAT2 | 0.646328 | 4 | 0 | 4 |
| B3GAT1 | KIF5A | 0.64442 | 5 | 0 | 5 |
| B3GAT1 | GABRB1 | 0.643431 | 3 | 0 | 3 |
| B3GAT1 | PDZD3 | 0.643385 | 5 | 0 | 5 |
| B3GAT1 | LCT | 0.643034 | 7 | 0 | 5 |
| B3GAT1 | CDH10 | 0.642652 | 6 | 0 | 5 |
| B3GAT1 | CNTD2 | 0.641822 | 7 | 0 | 6 |
| B3GAT1 | CER1 | 0.640392 | 4 | 0 | 4 |
| B3GAT1 | ADAM22 | 0.640307 | 5 | 0 | 4 |
For details and further investigation, click here