| Full name: FRAS1 related extracellular matrix 2 | Alias Symbol: DKFZp686J0811 | ||
| Type: protein-coding gene | Cytoband: 13q13.3 | ||
| Entrez ID: 341640 | HGNC ID: HGNC:25396 | Ensembl Gene: ENSG00000150893 | OMIM ID: 608945 |
| Drug and gene relationship at DGIdb | |||
Expression of FREM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FREM2 | 341640 | 230964_at | 0.4853 | 0.4315 | |
| GSE26886 | FREM2 | 341640 | 230964_at | 2.5198 | 0.0024 | |
| GSE45670 | FREM2 | 341640 | 230964_at | 0.9781 | 0.1947 | |
| GSE53622 | FREM2 | 341640 | 6867 | 0.7610 | 0.0000 | |
| GSE53624 | FREM2 | 341640 | 6867 | 0.7660 | 0.0000 | |
| GSE63941 | FREM2 | 341640 | 230964_at | 1.2680 | 0.3318 | |
| GSE77861 | FREM2 | 341640 | 230964_at | 0.8623 | 0.0792 | |
| SRP007169 | FREM2 | 341640 | RNAseq | 6.1406 | 0.0000 | |
| SRP133303 | FREM2 | 341640 | RNAseq | -0.0421 | 0.8449 | |
| SRP159526 | FREM2 | 341640 | RNAseq | 6.0807 | 0.0000 | |
| SRP193095 | FREM2 | 341640 | RNAseq | 2.3495 | 0.0000 | |
| TCGA | FREM2 | 341640 | RNAseq | -0.1683 | 0.7552 |
Upregulated datasets: 4; Downregulated datasets: 0.
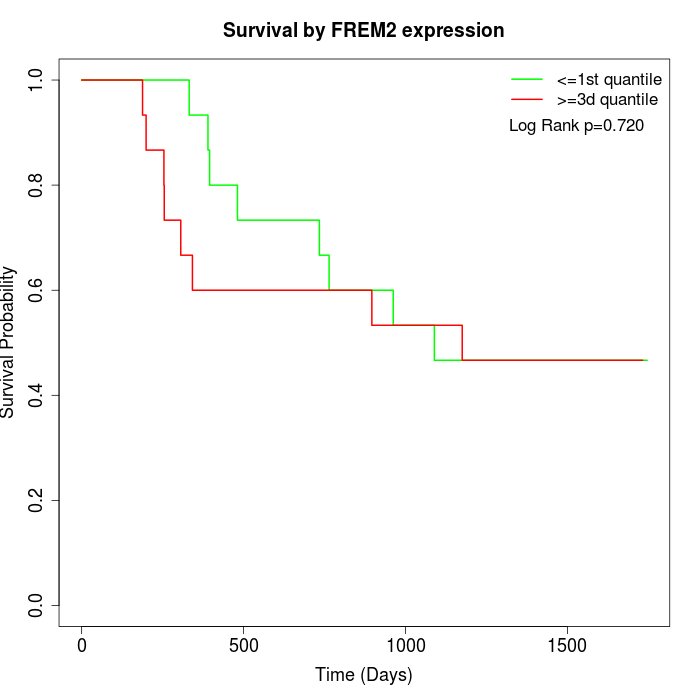
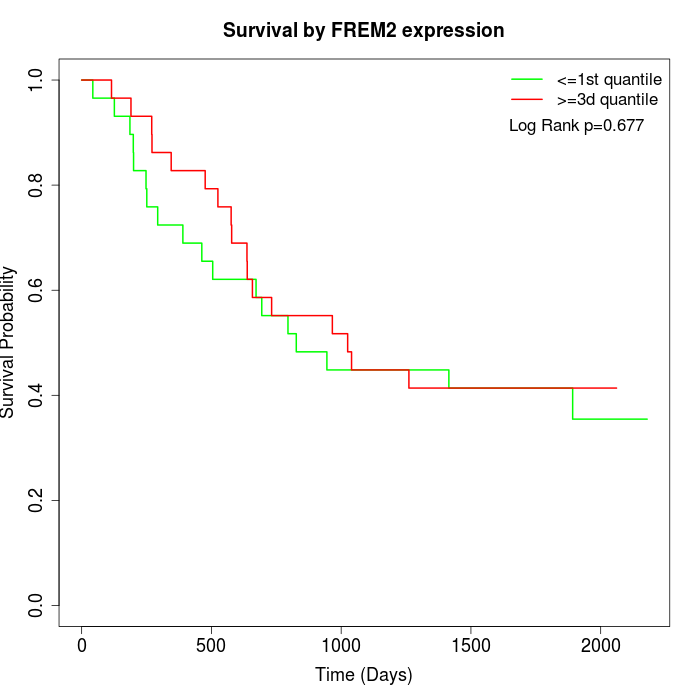
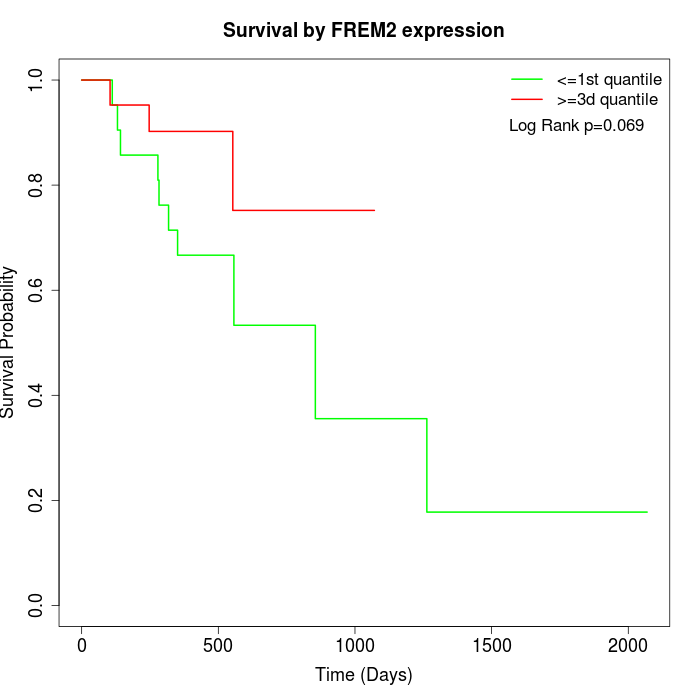
Survival by FREM2 expression:
Note: Click image to view full size file.
Copy number change of FREM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FREM2 | 341640 | 0 | 16 | 14 | |
| GSE20123 | FREM2 | 341640 | 0 | 15 | 15 | |
| GSE43470 | FREM2 | 341640 | 3 | 13 | 27 | |
| GSE46452 | FREM2 | 341640 | 0 | 33 | 26 | |
| GSE47630 | FREM2 | 341640 | 2 | 27 | 11 | |
| GSE54993 | FREM2 | 341640 | 12 | 2 | 56 | |
| GSE54994 | FREM2 | 341640 | 1 | 17 | 35 | |
| GSE60625 | FREM2 | 341640 | 0 | 3 | 8 | |
| GSE74703 | FREM2 | 341640 | 3 | 10 | 23 | |
| GSE74704 | FREM2 | 341640 | 0 | 12 | 8 | |
| TCGA | FREM2 | 341640 | 7 | 40 | 49 |
Total number of gains: 28; Total number of losses: 188; Total Number of normals: 272.
Somatic mutations of FREM2:
Generating mutation plots.
Highly correlated genes for FREM2:
Showing top 20/261 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FREM2 | CASC15 | 0.734809 | 4 | 0 | 3 |
| FREM2 | PRPSAP2 | 0.726196 | 3 | 0 | 3 |
| FREM2 | PLD3 | 0.718525 | 3 | 0 | 3 |
| FREM2 | RAP1GAP2 | 0.702131 | 3 | 0 | 3 |
| FREM2 | DLX6-AS1 | 0.685373 | 5 | 0 | 3 |
| FREM2 | JAKMIP3 | 0.68485 | 6 | 0 | 4 |
| FREM2 | TMEM163 | 0.681169 | 5 | 0 | 4 |
| FREM2 | CLDN20 | 0.676023 | 5 | 0 | 5 |
| FREM2 | KLHL22 | 0.668856 | 3 | 0 | 3 |
| FREM2 | FBN2 | 0.664625 | 6 | 0 | 4 |
| FREM2 | TBCE | 0.659905 | 3 | 0 | 3 |
| FREM2 | DDX10 | 0.65529 | 3 | 0 | 3 |
| FREM2 | ARTN | 0.653665 | 5 | 0 | 5 |
| FREM2 | AK8 | 0.652449 | 4 | 0 | 4 |
| FREM2 | SLC38A10 | 0.650343 | 4 | 0 | 3 |
| FREM2 | SCN9A | 0.649916 | 7 | 0 | 6 |
| FREM2 | DGCR5 | 0.644415 | 4 | 0 | 3 |
| FREM2 | AKR1B1 | 0.643202 | 4 | 0 | 4 |
| FREM2 | GLI2 | 0.63957 | 7 | 0 | 4 |
| FREM2 | PKN1 | 0.63612 | 4 | 0 | 3 |
For details and further investigation, click here