| Full name: kelch like family member 22 | Alias Symbol: FLJ14360|KELCHL | ||
| Type: protein-coding gene | Cytoband: 22q11.21 | ||
| Entrez ID: 84861 | HGNC ID: HGNC:25888 | Ensembl Gene: ENSG00000099910 | OMIM ID: 618020 |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL22 | 84861 | 49329_at | 0.2656 | 0.4082 | |
| GSE20347 | KLHL22 | 84861 | 49329_at | 0.2429 | 0.0389 | |
| GSE23400 | KLHL22 | 84861 | 49329_at | 0.0988 | 0.0412 | |
| GSE26886 | KLHL22 | 84861 | 49329_at | 1.3002 | 0.0000 | |
| GSE29001 | KLHL22 | 84861 | 49329_at | 0.3477 | 0.3030 | |
| GSE38129 | KLHL22 | 84861 | 49329_at | 0.1912 | 0.2529 | |
| GSE45670 | KLHL22 | 84861 | 49329_at | 0.2370 | 0.2692 | |
| GSE53622 | KLHL22 | 84861 | 9177 | 0.4361 | 0.0000 | |
| GSE53624 | KLHL22 | 84861 | 80416 | 0.5551 | 0.0000 | |
| GSE63941 | KLHL22 | 84861 | 49329_at | -0.6884 | 0.2723 | |
| GSE77861 | KLHL22 | 84861 | 49329_at | 0.4661 | 0.0495 | |
| GSE97050 | KLHL22 | 84861 | A_24_P117323 | -0.0831 | 0.7043 | |
| SRP007169 | KLHL22 | 84861 | RNAseq | 0.6171 | 0.1524 | |
| SRP008496 | KLHL22 | 84861 | RNAseq | 1.0793 | 0.0003 | |
| SRP064894 | KLHL22 | 84861 | RNAseq | 0.9383 | 0.0015 | |
| SRP133303 | KLHL22 | 84861 | RNAseq | 0.3378 | 0.0631 | |
| SRP159526 | KLHL22 | 84861 | RNAseq | 0.7701 | 0.0573 | |
| SRP193095 | KLHL22 | 84861 | RNAseq | 0.7991 | 0.0000 | |
| SRP219564 | KLHL22 | 84861 | RNAseq | 0.7976 | 0.1197 | |
| TCGA | KLHL22 | 84861 | RNAseq | -0.1151 | 0.0683 |
Upregulated datasets: 2; Downregulated datasets: 0.
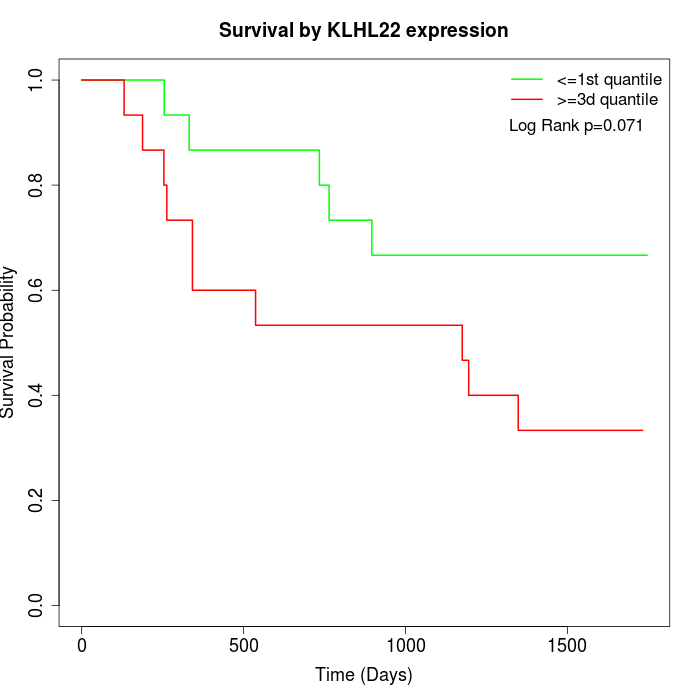
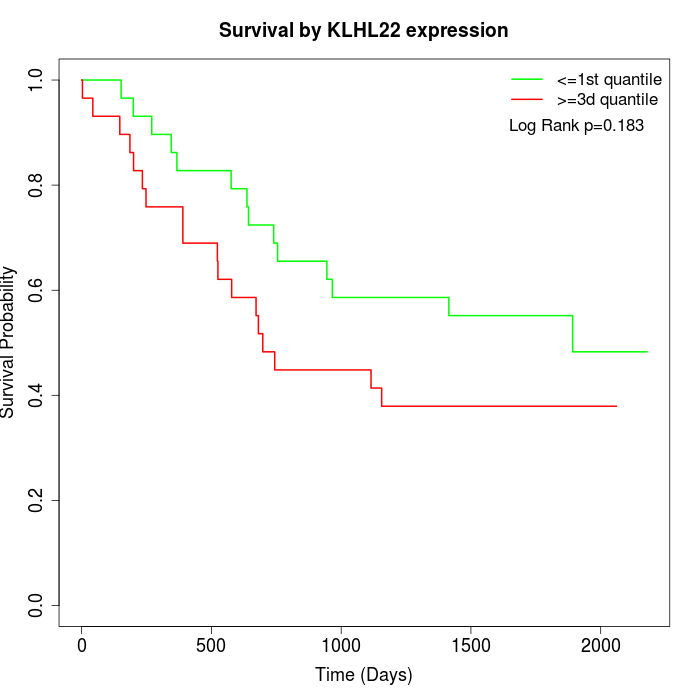
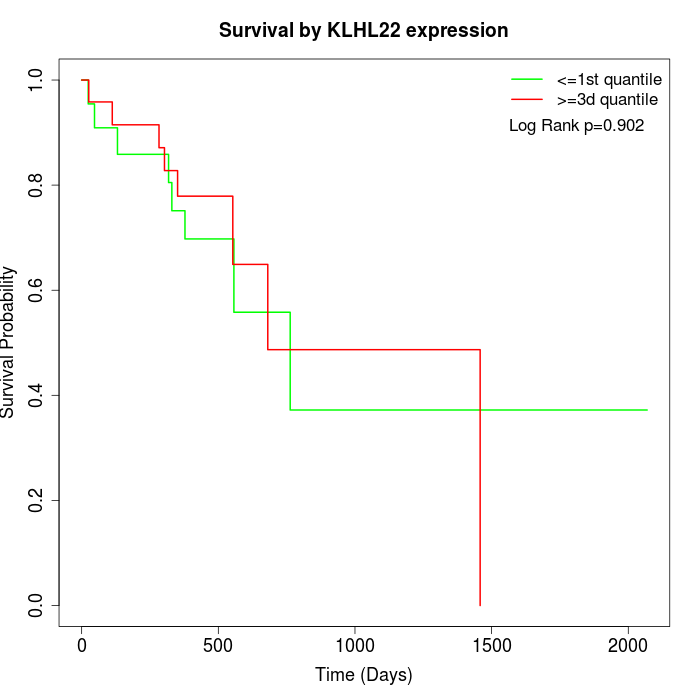
Survival by KLHL22 expression:
Note: Click image to view full size file.
Copy number change of KLHL22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL22 | 84861 | 5 | 7 | 18 | |
| GSE20123 | KLHL22 | 84861 | 5 | 7 | 18 | |
| GSE43470 | KLHL22 | 84861 | 4 | 7 | 32 | |
| GSE46452 | KLHL22 | 84861 | 33 | 1 | 25 | |
| GSE47630 | KLHL22 | 84861 | 8 | 5 | 27 | |
| GSE54993 | KLHL22 | 84861 | 3 | 6 | 61 | |
| GSE54994 | KLHL22 | 84861 | 11 | 7 | 35 | |
| GSE60625 | KLHL22 | 84861 | 5 | 0 | 6 | |
| GSE74703 | KLHL22 | 84861 | 4 | 5 | 27 | |
| GSE74704 | KLHL22 | 84861 | 3 | 4 | 13 | |
| TCGA | KLHL22 | 84861 | 27 | 17 | 52 |
Total number of gains: 108; Total number of losses: 66; Total Number of normals: 314.
Somatic mutations of KLHL22:
Generating mutation plots.
Highly correlated genes for KLHL22:
Showing top 20/457 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL22 | FGFBP2 | 0.706753 | 4 | 0 | 4 |
| KLHL22 | CCDC86 | 0.703676 | 3 | 0 | 3 |
| KLHL22 | SYCE1L | 0.688868 | 3 | 0 | 3 |
| KLHL22 | TSC22D1 | 0.687385 | 3 | 0 | 3 |
| KLHL22 | C6orf226 | 0.678987 | 3 | 0 | 3 |
| KLHL22 | SAC3D1 | 0.676621 | 5 | 0 | 5 |
| KLHL22 | TENM2 | 0.674201 | 3 | 0 | 3 |
| KLHL22 | MEF2C | 0.673773 | 3 | 0 | 3 |
| KLHL22 | LAMA3 | 0.672972 | 4 | 0 | 3 |
| KLHL22 | CASC15 | 0.669383 | 3 | 0 | 3 |
| KLHL22 | FREM2 | 0.668856 | 3 | 0 | 3 |
| KLHL22 | DVL2 | 0.668803 | 4 | 0 | 3 |
| KLHL22 | FBXO44 | 0.665603 | 4 | 0 | 4 |
| KLHL22 | TPM1 | 0.662897 | 4 | 0 | 4 |
| KLHL22 | GNAO1 | 0.662417 | 3 | 0 | 3 |
| KLHL22 | ANKRD52 | 0.653669 | 6 | 0 | 5 |
| KLHL22 | UGGT1 | 0.651782 | 4 | 0 | 4 |
| KLHL22 | MAP1B | 0.651779 | 4 | 0 | 3 |
| KLHL22 | RNASE1 | 0.651344 | 3 | 0 | 3 |
| KLHL22 | DCHS1 | 0.646841 | 4 | 0 | 4 |
For details and further investigation, click here