| Full name: glucagon receptor | Alias Symbol: GGR | ||
| Type: protein-coding gene | Cytoband: 17q25.3 | ||
| Entrez ID: 2642 | HGNC ID: HGNC:4192 | Ensembl Gene: ENSG00000215644 | OMIM ID: 138033 |
| Drug and gene relationship at DGIdb | |||
GCGR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04922 | Glucagon signaling pathway |
Expression of GCGR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GCGR | 2642 | 210565_at | 0.0226 | 0.9518 | |
| GSE20347 | GCGR | 2642 | 210565_at | 0.0742 | 0.4840 | |
| GSE23400 | GCGR | 2642 | 210565_at | -0.0264 | 0.5003 | |
| GSE26886 | GCGR | 2642 | 210565_at | -0.0761 | 0.5081 | |
| GSE29001 | GCGR | 2642 | 210565_at | -0.1393 | 0.3179 | |
| GSE38129 | GCGR | 2642 | 210565_at | 0.0149 | 0.8953 | |
| GSE45670 | GCGR | 2642 | 210565_at | 0.1483 | 0.0804 | |
| GSE53622 | GCGR | 2642 | 41630 | 0.4048 | 0.0198 | |
| GSE53624 | GCGR | 2642 | 41630 | 0.9424 | 0.0000 | |
| GSE63941 | GCGR | 2642 | 210565_at | 0.0564 | 0.7597 | |
| GSE77861 | GCGR | 2642 | 210565_at | 0.1065 | 0.3181 | |
| TCGA | GCGR | 2642 | RNAseq | 2.3789 | 0.0041 |
Upregulated datasets: 1; Downregulated datasets: 0.
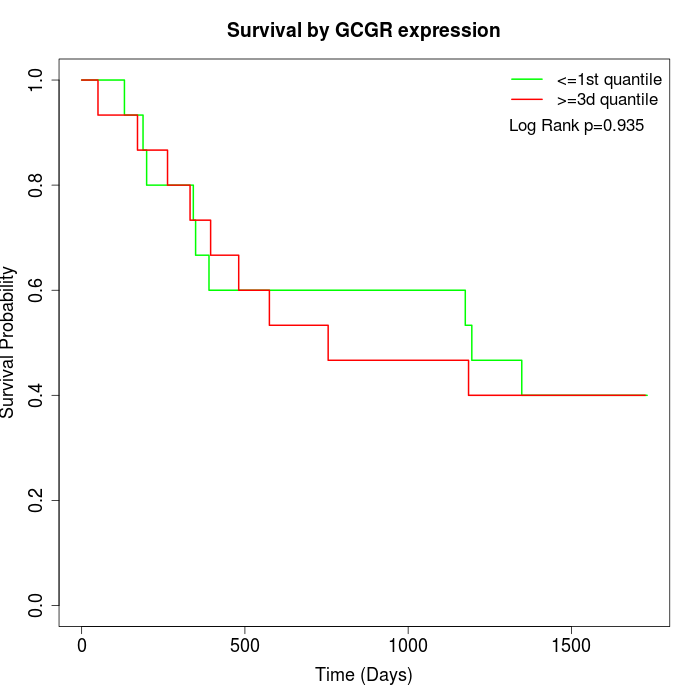
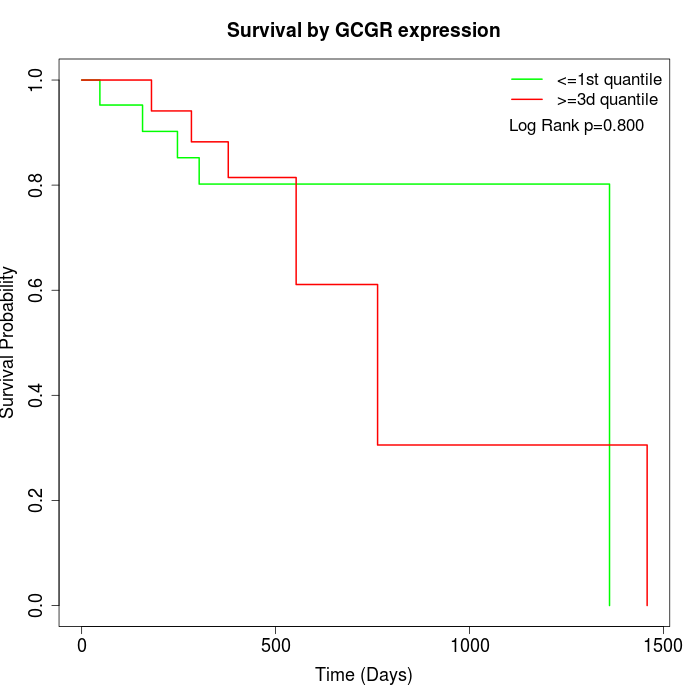
Survival by GCGR expression:
Note: Click image to view full size file.
Copy number change of GCGR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GCGR | 2642 | 4 | 4 | 22 | |
| GSE20123 | GCGR | 2642 | 4 | 4 | 22 | |
| GSE43470 | GCGR | 2642 | 3 | 5 | 35 | |
| GSE46452 | GCGR | 2642 | 34 | 0 | 25 | |
| GSE47630 | GCGR | 2642 | 9 | 2 | 29 | |
| GSE54993 | GCGR | 2642 | 2 | 5 | 63 | |
| GSE54994 | GCGR | 2642 | 10 | 6 | 37 | |
| GSE60625 | GCGR | 2642 | 6 | 0 | 5 | |
| GSE74703 | GCGR | 2642 | 3 | 3 | 30 | |
| GSE74704 | GCGR | 2642 | 4 | 3 | 13 | |
| TCGA | GCGR | 2642 | 25 | 12 | 59 |
Total number of gains: 104; Total number of losses: 44; Total Number of normals: 340.
Somatic mutations of GCGR:
Generating mutation plots.
Highly correlated genes for GCGR:
Showing top 20/308 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GCGR | HBQ1 | 0.669803 | 3 | 0 | 3 |
| GCGR | NR0B1 | 0.6557 | 3 | 0 | 3 |
| GCGR | GNAT1 | 0.637758 | 5 | 0 | 4 |
| GCGR | SEC14L4 | 0.633218 | 3 | 0 | 3 |
| GCGR | CSRP3 | 0.628347 | 4 | 0 | 4 |
| GCGR | GPR12 | 0.625311 | 4 | 0 | 4 |
| GCGR | OR2H2 | 0.623654 | 4 | 0 | 4 |
| GCGR | RNF185-AS1 | 0.622794 | 6 | 0 | 6 |
| GCGR | OR12D2 | 0.62166 | 5 | 0 | 4 |
| GCGR | ADAMTS7 | 0.620947 | 3 | 0 | 3 |
| GCGR | CXCR1 | 0.614262 | 5 | 0 | 4 |
| GCGR | CPA2 | 0.611005 | 5 | 0 | 5 |
| GCGR | PPP1R14D | 0.610756 | 4 | 0 | 4 |
| GCGR | GUCA2B | 0.608549 | 5 | 0 | 4 |
| GCGR | COL11A2 | 0.604949 | 6 | 0 | 5 |
| GCGR | TLE6 | 0.604125 | 4 | 0 | 4 |
| GCGR | GRK1 | 0.603368 | 4 | 0 | 4 |
| GCGR | PAPPA2 | 0.598758 | 7 | 0 | 6 |
| GCGR | GRM6 | 0.598164 | 4 | 0 | 4 |
| GCGR | SHBG | 0.597693 | 5 | 0 | 4 |
For details and further investigation, click here