| Full name: carboxypeptidase A2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q32.2 | ||
| Entrez ID: 1358 | HGNC ID: HGNC:2297 | Ensembl Gene: ENSG00000158516 | OMIM ID: 600688 |
| Drug and gene relationship at DGIdb | |||
Expression of CPA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CPA2 | 1358 | 206212_at | 0.0154 | 0.9764 | |
| GSE20347 | CPA2 | 1358 | 206212_at | -0.0763 | 0.5175 | |
| GSE23400 | CPA2 | 1358 | 206212_at | -0.1685 | 0.0018 | |
| GSE26886 | CPA2 | 1358 | 206212_at | -0.0524 | 0.6796 | |
| GSE29001 | CPA2 | 1358 | 206212_at | -0.1970 | 0.2076 | |
| GSE38129 | CPA2 | 1358 | 206212_at | -0.4156 | 0.0897 | |
| GSE45670 | CPA2 | 1358 | 206212_at | 0.0877 | 0.3344 | |
| GSE53622 | CPA2 | 1358 | 16958 | -0.0222 | 0.9498 | |
| GSE53624 | CPA2 | 1358 | 16958 | 0.0510 | 0.8682 | |
| GSE63941 | CPA2 | 1358 | 206212_at | -0.0940 | 0.5789 | |
| GSE77861 | CPA2 | 1358 | 206212_at | -0.0207 | 0.8599 | |
| TCGA | CPA2 | 1358 | RNAseq | -5.2920 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
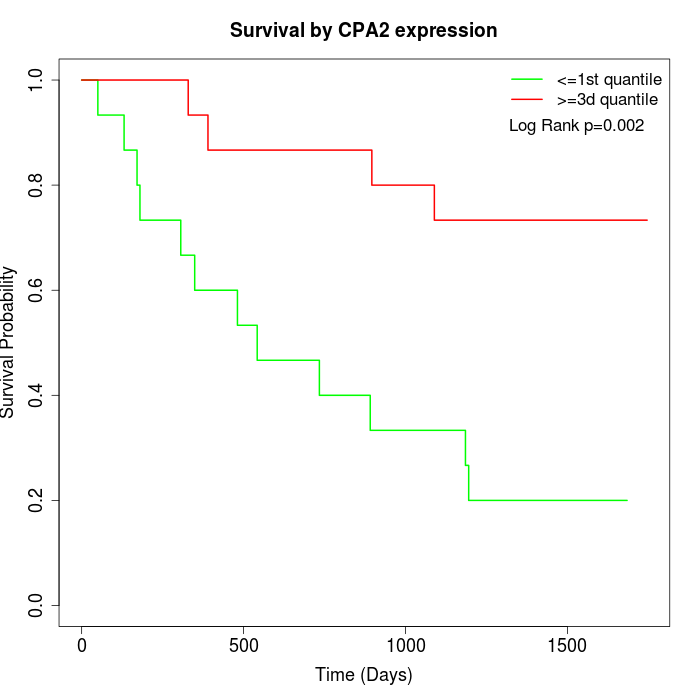
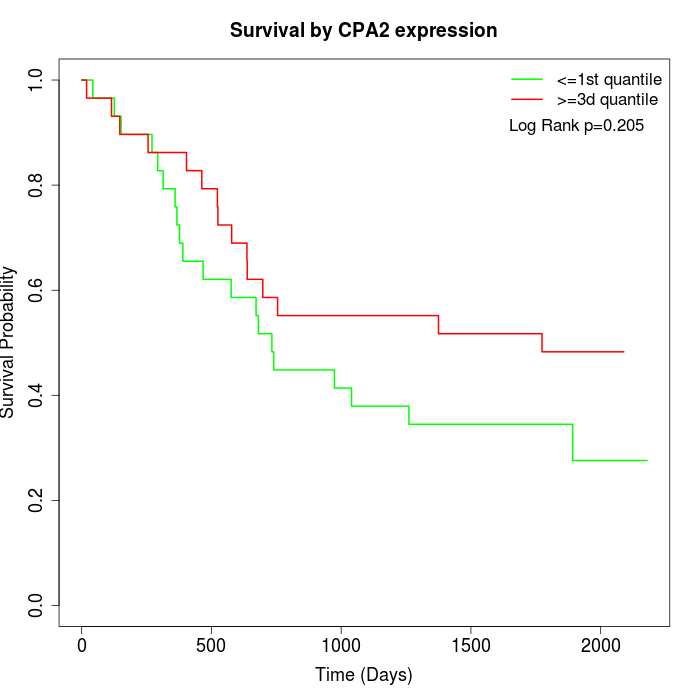
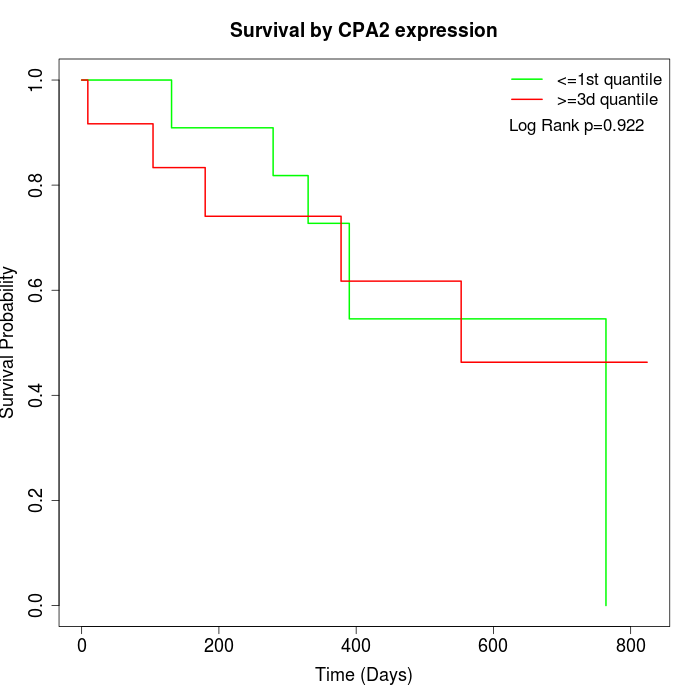
Survival by CPA2 expression:
Note: Click image to view full size file.
Copy number change of CPA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CPA2 | 1358 | 8 | 1 | 21 | |
| GSE20123 | CPA2 | 1358 | 8 | 1 | 21 | |
| GSE43470 | CPA2 | 1358 | 2 | 3 | 38 | |
| GSE46452 | CPA2 | 1358 | 7 | 2 | 50 | |
| GSE47630 | CPA2 | 1358 | 7 | 4 | 29 | |
| GSE54993 | CPA2 | 1358 | 3 | 5 | 62 | |
| GSE54994 | CPA2 | 1358 | 7 | 7 | 39 | |
| GSE60625 | CPA2 | 1358 | 0 | 0 | 11 | |
| GSE74703 | CPA2 | 1358 | 2 | 3 | 31 | |
| GSE74704 | CPA2 | 1358 | 4 | 1 | 15 | |
| TCGA | CPA2 | 1358 | 29 | 21 | 46 |
Total number of gains: 77; Total number of losses: 48; Total Number of normals: 363.
Somatic mutations of CPA2:
Generating mutation plots.
Highly correlated genes for CPA2:
Showing top 20/658 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CPA2 | KRTAP5-8 | 0.799639 | 3 | 0 | 3 |
| CPA2 | MTAP | 0.785181 | 3 | 0 | 3 |
| CPA2 | RNF17 | 0.763771 | 3 | 0 | 3 |
| CPA2 | GC | 0.753265 | 5 | 0 | 5 |
| CPA2 | ZNF460 | 0.749592 | 3 | 0 | 3 |
| CPA2 | TULP2 | 0.743374 | 3 | 0 | 3 |
| CPA2 | KSR1 | 0.740359 | 3 | 0 | 3 |
| CPA2 | USP2 | 0.736608 | 3 | 0 | 3 |
| CPA2 | ATP4A | 0.734944 | 6 | 0 | 5 |
| CPA2 | IL18BP | 0.734574 | 3 | 0 | 3 |
| CPA2 | CELA3A | 0.733871 | 5 | 0 | 5 |
| CPA2 | CRISP1 | 0.73277 | 3 | 0 | 3 |
| CPA2 | CHAT | 0.725816 | 3 | 0 | 3 |
| CPA2 | KIR3DL3 | 0.72487 | 3 | 0 | 3 |
| CPA2 | GPR15 | 0.724663 | 3 | 0 | 3 |
| CPA2 | CRP | 0.724304 | 3 | 0 | 3 |
| CPA2 | COX6A2 | 0.721905 | 3 | 0 | 3 |
| CPA2 | CDH7 | 0.720491 | 3 | 0 | 3 |
| CPA2 | SCNN1G | 0.718179 | 3 | 0 | 3 |
| CPA2 | DAB1 | 0.715625 | 4 | 0 | 4 |
For details and further investigation, click here