| Full name: gap junction protein alpha 1 | Alias Symbol: CX43|ODD|ODOD|SDTY3 | ||
| Type: protein-coding gene | Cytoband: 6q22.31 | ||
| Entrez ID: 2697 | HGNC ID: HGNC:4274 | Ensembl Gene: ENSG00000152661 | OMIM ID: 121014 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
GJA1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04540 | Gap junction |
Expression of GJA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GJA1 | 2697 | 201667_at | -0.0699 | 0.9445 | |
| GSE20347 | GJA1 | 2697 | 201667_at | 0.1356 | 0.6073 | |
| GSE23400 | GJA1 | 2697 | 201667_at | 0.5742 | 0.0004 | |
| GSE26886 | GJA1 | 2697 | 201667_at | 0.5947 | 0.0274 | |
| GSE29001 | GJA1 | 2697 | 201667_at | 0.9337 | 0.0456 | |
| GSE38129 | GJA1 | 2697 | 201667_at | 0.1585 | 0.4135 | |
| GSE45670 | GJA1 | 2697 | 201667_at | 0.4690 | 0.0385 | |
| GSE53622 | GJA1 | 2697 | 23360 | 0.7348 | 0.0000 | |
| GSE53624 | GJA1 | 2697 | 23360 | 0.8254 | 0.0000 | |
| GSE63941 | GJA1 | 2697 | 201667_at | -2.6913 | 0.3031 | |
| GSE77861 | GJA1 | 2697 | 201667_at | 0.5959 | 0.1510 | |
| GSE97050 | GJA1 | 2697 | A_24_P55295 | -0.0056 | 0.9930 | |
| SRP007169 | GJA1 | 2697 | RNAseq | 0.1700 | 0.6939 | |
| SRP008496 | GJA1 | 2697 | RNAseq | 1.1240 | 0.0078 | |
| SRP064894 | GJA1 | 2697 | RNAseq | 0.6397 | 0.0416 | |
| SRP133303 | GJA1 | 2697 | RNAseq | 1.3586 | 0.0000 | |
| SRP159526 | GJA1 | 2697 | RNAseq | 0.3758 | 0.4801 | |
| SRP193095 | GJA1 | 2697 | RNAseq | 0.5783 | 0.0690 | |
| SRP219564 | GJA1 | 2697 | RNAseq | -0.0182 | 0.9808 | |
| TCGA | GJA1 | 2697 | RNAseq | 0.3816 | 0.0004 |
Upregulated datasets: 2; Downregulated datasets: 0.
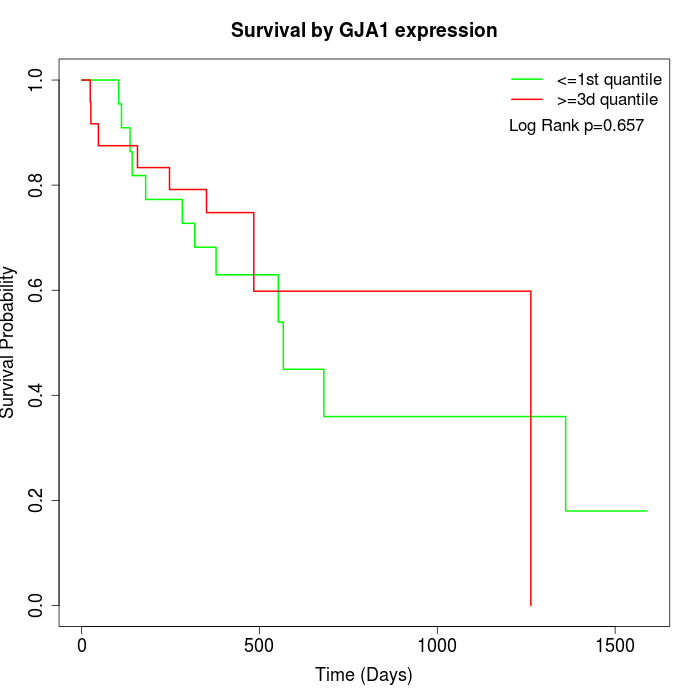
Survival by GJA1 expression:
Note: Click image to view full size file.
Copy number change of GJA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GJA1 | 2697 | 1 | 4 | 25 | |
| GSE20123 | GJA1 | 2697 | 1 | 3 | 26 | |
| GSE43470 | GJA1 | 2697 | 4 | 1 | 38 | |
| GSE46452 | GJA1 | 2697 | 2 | 11 | 46 | |
| GSE47630 | GJA1 | 2697 | 9 | 4 | 27 | |
| GSE54993 | GJA1 | 2697 | 3 | 2 | 65 | |
| GSE54994 | GJA1 | 2697 | 8 | 8 | 37 | |
| GSE60625 | GJA1 | 2697 | 0 | 1 | 10 | |
| GSE74703 | GJA1 | 2697 | 3 | 1 | 32 | |
| GSE74704 | GJA1 | 2697 | 0 | 1 | 19 | |
| TCGA | GJA1 | 2697 | 9 | 19 | 68 |
Total number of gains: 40; Total number of losses: 55; Total Number of normals: 393.
Somatic mutations of GJA1:
Generating mutation plots.
Highly correlated genes for GJA1:
Showing top 20/256 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GJA1 | PHLDA2 | 0.712281 | 3 | 0 | 3 |
| GJA1 | JDP2 | 0.709387 | 3 | 0 | 3 |
| GJA1 | ARHGEF40 | 0.663307 | 3 | 0 | 3 |
| GJA1 | EGR1 | 0.654058 | 3 | 0 | 3 |
| GJA1 | COL9A3 | 0.651609 | 3 | 0 | 3 |
| GJA1 | MRPL45 | 0.651189 | 3 | 0 | 3 |
| GJA1 | JMY | 0.650208 | 3 | 0 | 3 |
| GJA1 | TENM2 | 0.639633 | 7 | 0 | 5 |
| GJA1 | ZC3H14 | 0.626228 | 4 | 0 | 3 |
| GJA1 | TINF2 | 0.625337 | 3 | 0 | 3 |
| GJA1 | DIAPH2 | 0.623386 | 3 | 0 | 3 |
| GJA1 | RNF145 | 0.615236 | 4 | 0 | 3 |
| GJA1 | G3BP1 | 0.614164 | 3 | 0 | 3 |
| GJA1 | KLF10 | 0.609685 | 10 | 0 | 7 |
| GJA1 | HELZ | 0.609425 | 3 | 0 | 3 |
| GJA1 | GRB10 | 0.606434 | 3 | 0 | 3 |
| GJA1 | MB21D2 | 0.604478 | 7 | 0 | 5 |
| GJA1 | BLACAT1 | 0.596055 | 3 | 0 | 3 |
| GJA1 | CRIPT | 0.59218 | 4 | 0 | 3 |
| GJA1 | ANXA4 | 0.591562 | 5 | 0 | 4 |
For details and further investigation, click here