| Full name: early growth response 1 | Alias Symbol: TIS8|G0S30|NGFI-A|KROX-24|ZIF-268|AT225|ZNF225 | ||
| Type: protein-coding gene | Cytoband: 5q31.2 | ||
| Entrez ID: 1958 | HGNC ID: HGNC:3238 | Ensembl Gene: ENSG00000120738 | OMIM ID: 128990 |
| Drug and gene relationship at DGIdb | |||
EGR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | |
| hsa05020 | Prion diseases |
Expression of EGR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGR1 | 1958 | 227404_s_at | -1.2711 | 0.2055 | |
| GSE20347 | EGR1 | 1958 | 201694_s_at | -0.2875 | 0.4148 | |
| GSE23400 | EGR1 | 1958 | 201694_s_at | -0.4657 | 0.0011 | |
| GSE26886 | EGR1 | 1958 | 227404_s_at | 1.4475 | 0.0277 | |
| GSE29001 | EGR1 | 1958 | 201694_s_at | -0.7919 | 0.0487 | |
| GSE38129 | EGR1 | 1958 | 201694_s_at | -0.6316 | 0.0544 | |
| GSE45670 | EGR1 | 1958 | 201694_s_at | -1.0193 | 0.0350 | |
| GSE53622 | EGR1 | 1958 | 168876 | -1.4223 | 0.0000 | |
| GSE53624 | EGR1 | 1958 | 168876 | -0.8735 | 0.0000 | |
| GSE63941 | EGR1 | 1958 | 227404_s_at | 3.5614 | 0.0037 | |
| GSE77861 | EGR1 | 1958 | 227404_s_at | 0.1020 | 0.9204 | |
| GSE97050 | EGR1 | 1958 | A_23_P214080 | -0.9979 | 0.2372 | |
| SRP007169 | EGR1 | 1958 | RNAseq | -0.3925 | 0.4190 | |
| SRP008496 | EGR1 | 1958 | RNAseq | -0.4110 | 0.4031 | |
| SRP064894 | EGR1 | 1958 | RNAseq | -0.6998 | 0.0655 | |
| SRP133303 | EGR1 | 1958 | RNAseq | -0.2285 | 0.5950 | |
| SRP159526 | EGR1 | 1958 | RNAseq | 0.4302 | 0.5213 | |
| SRP193095 | EGR1 | 1958 | RNAseq | 0.2672 | 0.5351 | |
| SRP219564 | EGR1 | 1958 | RNAseq | -1.0400 | 0.2615 | |
| TCGA | EGR1 | 1958 | RNAseq | -0.1396 | 0.1429 |
Upregulated datasets: 2; Downregulated datasets: 2.
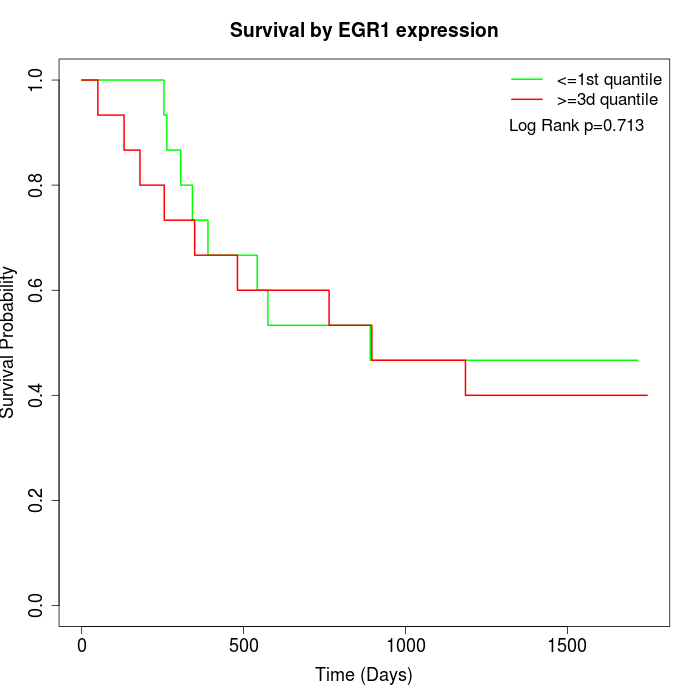
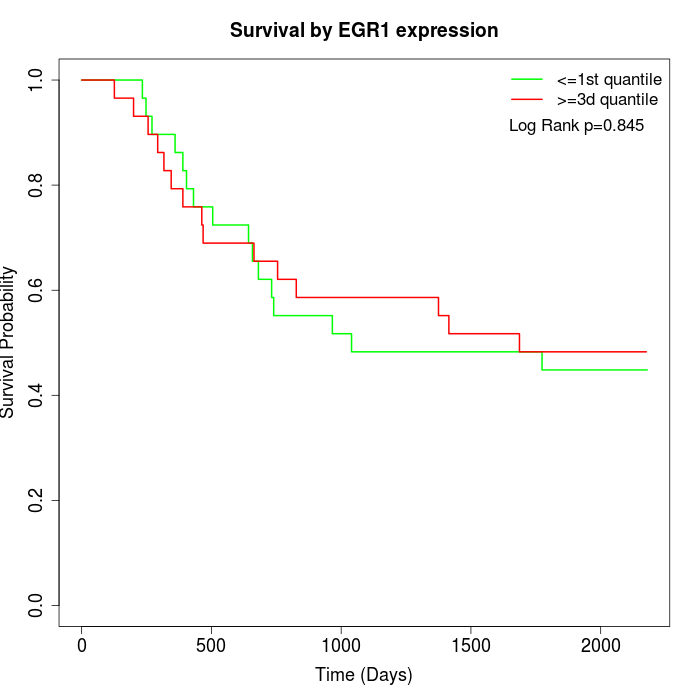
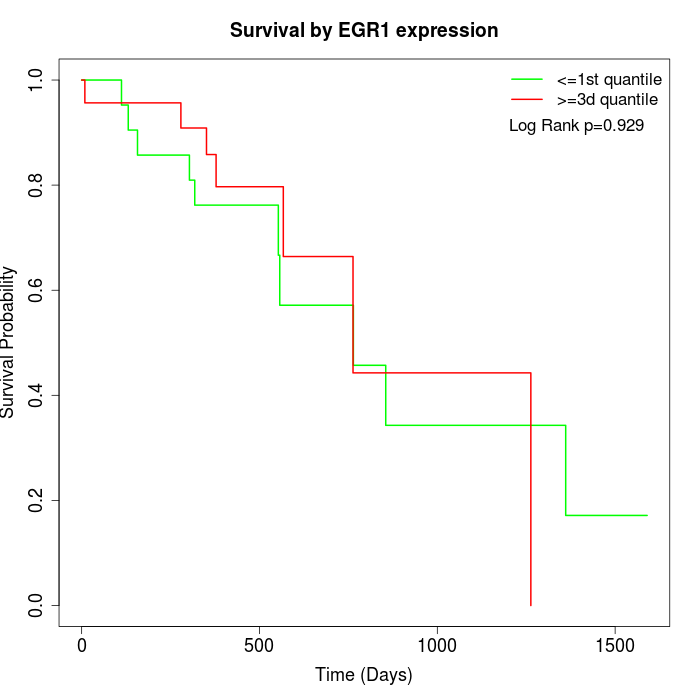
Survival by EGR1 expression:
Note: Click image to view full size file.
Copy number change of EGR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EGR1 | 1958 | 3 | 10 | 17 | |
| GSE20123 | EGR1 | 1958 | 4 | 10 | 16 | |
| GSE43470 | EGR1 | 1958 | 4 | 5 | 34 | |
| GSE46452 | EGR1 | 1958 | 0 | 27 | 32 | |
| GSE47630 | EGR1 | 1958 | 0 | 21 | 19 | |
| GSE54993 | EGR1 | 1958 | 9 | 1 | 60 | |
| GSE54994 | EGR1 | 1958 | 3 | 13 | 37 | |
| GSE60625 | EGR1 | 1958 | 0 | 0 | 11 | |
| GSE74703 | EGR1 | 1958 | 3 | 4 | 29 | |
| GSE74704 | EGR1 | 1958 | 3 | 4 | 13 | |
| TCGA | EGR1 | 1958 | 3 | 37 | 56 |
Total number of gains: 32; Total number of losses: 132; Total Number of normals: 324.
Somatic mutations of EGR1:
Generating mutation plots.
Highly correlated genes for EGR1:
Showing top 20/148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGR1 | FOS | 0.734328 | 13 | 0 | 11 |
| EGR1 | JUN | 0.71493 | 11 | 0 | 10 |
| EGR1 | APOBEC2 | 0.68773 | 3 | 0 | 3 |
| EGR1 | MIDN | 0.679539 | 3 | 0 | 3 |
| EGR1 | FOSB | 0.678116 | 13 | 0 | 11 |
| EGR1 | DUSP1 | 0.675648 | 12 | 0 | 10 |
| EGR1 | PTEN | 0.670025 | 3 | 0 | 3 |
| EGR1 | GRPR | 0.667191 | 3 | 0 | 3 |
| EGR1 | GJA1 | 0.654058 | 3 | 0 | 3 |
| EGR1 | JUNB | 0.652571 | 11 | 0 | 11 |
| EGR1 | C5orf24 | 0.646386 | 3 | 0 | 3 |
| EGR1 | MED7 | 0.644994 | 3 | 0 | 3 |
| EGR1 | IAH1 | 0.643313 | 3 | 0 | 3 |
| EGR1 | CBX4 | 0.640495 | 3 | 0 | 3 |
| EGR1 | ANKRA2 | 0.640238 | 4 | 0 | 3 |
| EGR1 | TPSD1 | 0.640221 | 3 | 0 | 3 |
| EGR1 | RSL24D1 | 0.637573 | 3 | 0 | 3 |
| EGR1 | ADAMTS10 | 0.635514 | 4 | 0 | 3 |
| EGR1 | IER2 | 0.629594 | 8 | 0 | 7 |
| EGR1 | PAIP2 | 0.628557 | 3 | 0 | 3 |
For details and further investigation, click here