| Full name: TERF1 interacting nuclear factor 2 | Alias Symbol: TIN2 | ||
| Type: protein-coding gene | Cytoband: 14q12 | ||
| Entrez ID: 26277 | HGNC ID: HGNC:11824 | Ensembl Gene: | OMIM ID: 604319 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TINF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TINF2 | 26277 | 224809_x_at | -0.2667 | 0.6279 | |
| GSE20347 | TINF2 | 26277 | 220052_s_at | 0.0877 | 0.5520 | |
| GSE23400 | TINF2 | 26277 | 220052_s_at | 0.0328 | 0.5662 | |
| GSE26886 | TINF2 | 26277 | 224809_x_at | 0.1348 | 0.5022 | |
| GSE29001 | TINF2 | 26277 | 220052_s_at | 0.0103 | 0.9617 | |
| GSE38129 | TINF2 | 26277 | 220052_s_at | 0.0017 | 0.9902 | |
| GSE45670 | TINF2 | 26277 | 224809_x_at | 0.0664 | 0.7148 | |
| GSE53622 | TINF2 | 26277 | 79997 | 0.0415 | 0.6411 | |
| GSE53624 | TINF2 | 26277 | 79997 | -0.0867 | 0.3614 | |
| GSE63941 | TINF2 | 26277 | 224809_x_at | 0.0564 | 0.9250 | |
| GSE77861 | TINF2 | 26277 | 224809_x_at | 0.0215 | 0.9590 | |
| GSE97050 | TINF2 | 26277 | A_33_P3372941 | 0.3644 | 0.2018 | |
| SRP007169 | TINF2 | 26277 | RNAseq | -0.8551 | 0.0227 | |
| SRP008496 | TINF2 | 26277 | RNAseq | -0.7658 | 0.0028 | |
| SRP064894 | TINF2 | 26277 | RNAseq | -0.1626 | 0.2439 | |
| SRP133303 | TINF2 | 26277 | RNAseq | -0.1089 | 0.6784 | |
| SRP159526 | TINF2 | 26277 | RNAseq | -0.5725 | 0.0075 | |
| SRP193095 | TINF2 | 26277 | RNAseq | -0.3047 | 0.0166 | |
| SRP219564 | TINF2 | 26277 | RNAseq | -0.6795 | 0.0109 | |
| TCGA | TINF2 | 26277 | RNAseq | 0.0074 | 0.8835 |
Upregulated datasets: 0; Downregulated datasets: 0.
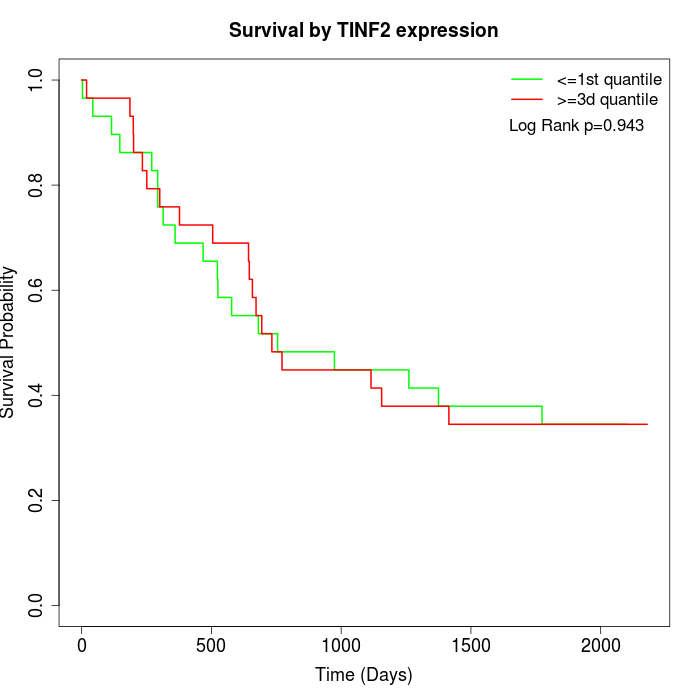
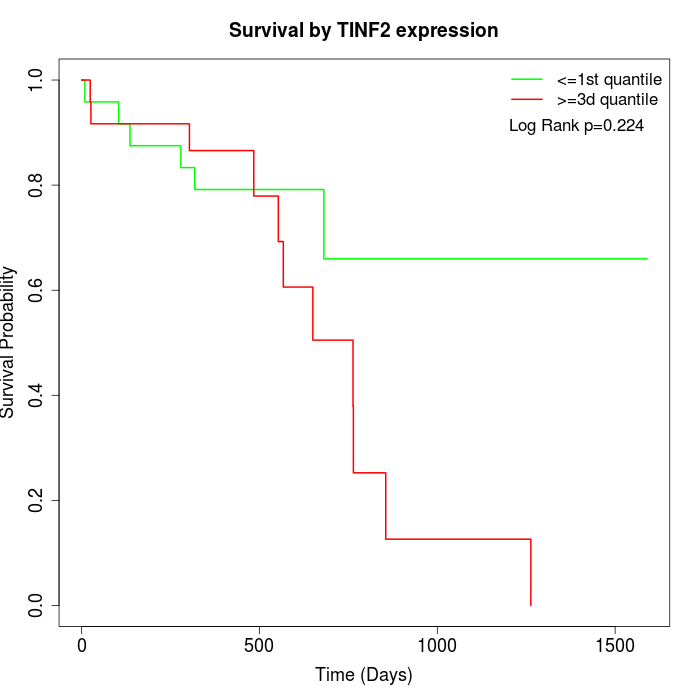
Survival by TINF2 expression:
Note: Click image to view full size file.
Copy number change of TINF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TINF2 | 26277 | 8 | 3 | 19 | |
| GSE20123 | TINF2 | 26277 | 8 | 3 | 19 | |
| GSE43470 | TINF2 | 26277 | 8 | 2 | 33 | |
| GSE46452 | TINF2 | 26277 | 16 | 2 | 41 | |
| GSE47630 | TINF2 | 26277 | 11 | 10 | 19 | |
| GSE54993 | TINF2 | 26277 | 3 | 11 | 56 | |
| GSE54994 | TINF2 | 26277 | 18 | 5 | 30 | |
| GSE60625 | TINF2 | 26277 | 0 | 2 | 9 | |
| GSE74703 | TINF2 | 26277 | 7 | 2 | 27 | |
| GSE74704 | TINF2 | 26277 | 3 | 2 | 15 | |
| TCGA | TINF2 | 26277 | 27 | 12 | 57 |
Total number of gains: 109; Total number of losses: 54; Total Number of normals: 325.
Somatic mutations of TINF2:
Generating mutation plots.
Highly correlated genes for TINF2:
Showing top 20/136 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TINF2 | RASA1 | 0.729761 | 3 | 0 | 3 |
| TINF2 | RAD50 | 0.72665 | 3 | 0 | 3 |
| TINF2 | PROS1 | 0.72535 | 3 | 0 | 3 |
| TINF2 | JOSD2 | 0.7246 | 3 | 0 | 3 |
| TINF2 | C16orf87 | 0.724478 | 3 | 0 | 3 |
| TINF2 | ALDOA | 0.720628 | 3 | 0 | 3 |
| TINF2 | HDAC3 | 0.699858 | 4 | 0 | 3 |
| TINF2 | RAB12 | 0.698173 | 3 | 0 | 3 |
| TINF2 | HSPA4 | 0.697085 | 3 | 0 | 3 |
| TINF2 | S1PR5 | 0.691055 | 3 | 0 | 3 |
| TINF2 | FHL2 | 0.68738 | 4 | 0 | 4 |
| TINF2 | EAF1 | 0.685744 | 3 | 0 | 3 |
| TINF2 | CAPN3 | 0.684612 | 3 | 0 | 3 |
| TINF2 | FNIP1 | 0.68402 | 3 | 0 | 3 |
| TINF2 | PPP6C | 0.682616 | 3 | 0 | 3 |
| TINF2 | VTI1B | 0.680818 | 5 | 0 | 4 |
| TINF2 | IRS1 | 0.676588 | 3 | 0 | 3 |
| TINF2 | MED7 | 0.672095 | 3 | 0 | 3 |
| TINF2 | SLC6A2 | 0.66655 | 3 | 0 | 3 |
| TINF2 | SLU7 | 0.663512 | 4 | 0 | 3 |
For details and further investigation, click here