| Full name: glycine-N-acyltransferase like 2 | Alias Symbol: BXMAS2-10|MGC24009 | ||
| Type: protein-coding gene | Cytoband: 11q12.1 | ||
| Entrez ID: 219970 | HGNC ID: HGNC:24178 | Ensembl Gene: ENSG00000156689 | OMIM ID: 614762 |
| Drug and gene relationship at DGIdb | |||
Expression of GLYATL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLYATL2 | 219970 | 1554712_a_at | 0.0984 | 0.9234 | |
| GSE26886 | GLYATL2 | 219970 | 1554712_a_at | 0.0524 | 0.6289 | |
| GSE45670 | GLYATL2 | 219970 | 1554712_a_at | -0.2781 | 0.0935 | |
| GSE53622 | GLYATL2 | 219970 | 56982 | -1.1493 | 0.0000 | |
| GSE53624 | GLYATL2 | 219970 | 56982 | -1.1709 | 0.0000 | |
| GSE63941 | GLYATL2 | 219970 | 1554712_a_at | 0.0473 | 0.7859 | |
| GSE77861 | GLYATL2 | 219970 | 1554712_a_at | 0.0662 | 0.4335 | |
| GSE97050 | GLYATL2 | 219970 | A_23_P47484 | 0.2092 | 0.4191 | |
| SRP133303 | GLYATL2 | 219970 | RNAseq | -0.9099 | 0.2162 | |
| TCGA | GLYATL2 | 219970 | RNAseq | 0.8264 | 0.2030 |
Upregulated datasets: 0; Downregulated datasets: 2.
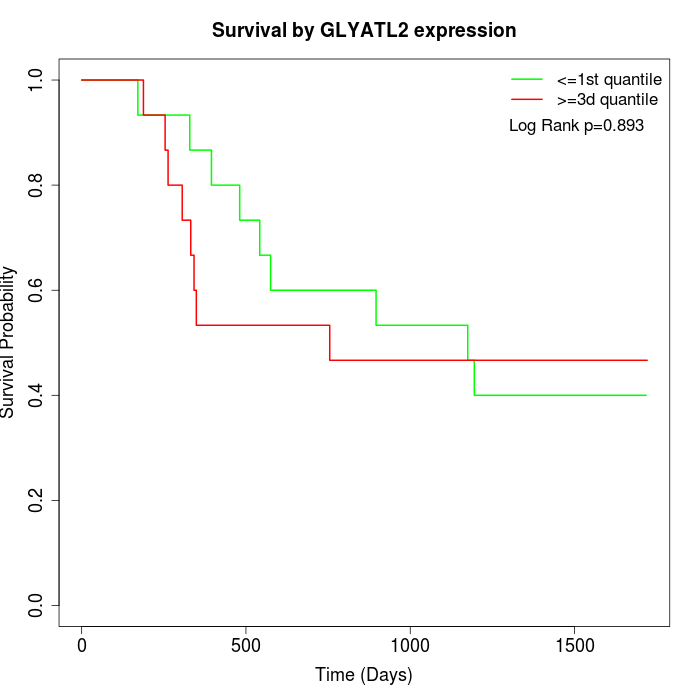
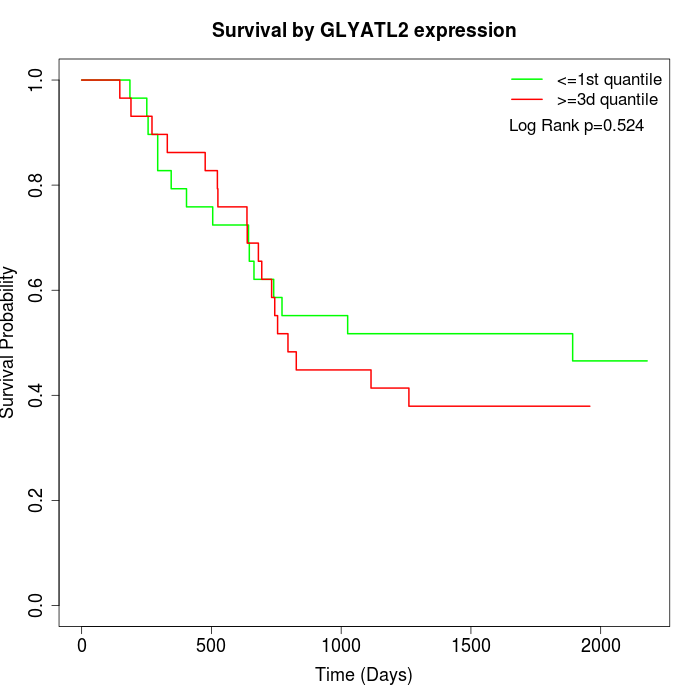
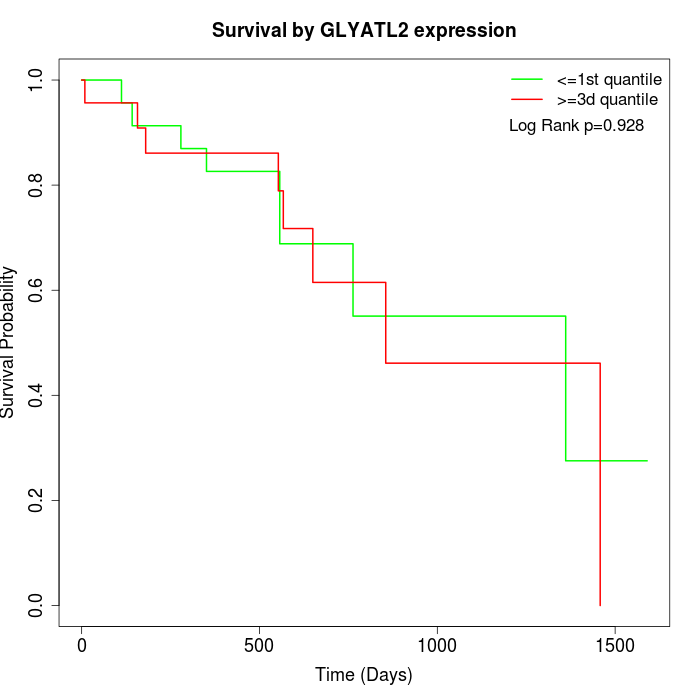
Survival by GLYATL2 expression:
Note: Click image to view full size file.
Copy number change of GLYATL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLYATL2 | 219970 | 1 | 4 | 25 | |
| GSE20123 | GLYATL2 | 219970 | 1 | 4 | 25 | |
| GSE43470 | GLYATL2 | 219970 | 1 | 5 | 37 | |
| GSE46452 | GLYATL2 | 219970 | 10 | 4 | 45 | |
| GSE47630 | GLYATL2 | 219970 | 3 | 8 | 29 | |
| GSE54993 | GLYATL2 | 219970 | 3 | 0 | 67 | |
| GSE54994 | GLYATL2 | 219970 | 5 | 6 | 42 | |
| GSE60625 | GLYATL2 | 219970 | 0 | 3 | 8 | |
| GSE74703 | GLYATL2 | 219970 | 1 | 3 | 32 | |
| GSE74704 | GLYATL2 | 219970 | 1 | 4 | 15 | |
| TCGA | GLYATL2 | 219970 | 14 | 10 | 72 |
Total number of gains: 40; Total number of losses: 51; Total Number of normals: 397.
Somatic mutations of GLYATL2:
Generating mutation plots.
Highly correlated genes for GLYATL2:
Showing top 20/37 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLYATL2 | NXPE4 | 0.764599 | 4 | 0 | 4 |
| GLYATL2 | BPIFB2 | 0.690808 | 4 | 0 | 4 |
| GLYATL2 | TFF3 | 0.686576 | 4 | 0 | 4 |
| GLYATL2 | BPIFB6 | 0.68016 | 4 | 0 | 4 |
| GLYATL2 | MSMB | 0.67048 | 3 | 0 | 3 |
| GLYATL2 | BPIFB1 | 0.659046 | 4 | 0 | 4 |
| GLYATL2 | TSPAN8 | 0.653927 | 4 | 0 | 4 |
| GLYATL2 | MUC5B | 0.644688 | 4 | 0 | 4 |
| GLYATL2 | LRRC26 | 0.641254 | 3 | 0 | 3 |
| GLYATL2 | WFDC2 | 0.635807 | 5 | 0 | 5 |
| GLYATL2 | CHST9 | 0.615353 | 3 | 0 | 3 |
| GLYATL2 | KIAA1324 | 0.613736 | 5 | 0 | 4 |
| GLYATL2 | PROM1 | 0.609779 | 4 | 0 | 4 |
| GLYATL2 | PIP | 0.607979 | 4 | 0 | 3 |
| GLYATL2 | TMEM211 | 0.60528 | 3 | 0 | 3 |
| GLYATL2 | SCGB2A2 | 0.604681 | 4 | 0 | 3 |
| GLYATL2 | NKX3-1 | 0.604668 | 5 | 0 | 4 |
| GLYATL2 | PIGR | 0.597148 | 3 | 0 | 3 |
| GLYATL2 | ZG16B | 0.593042 | 5 | 0 | 4 |
| GLYATL2 | C6orf58 | 0.590273 | 5 | 0 | 3 |
For details and further investigation, click here