| Full name: G protein nucleolar 2 | Alias Symbol: Ngp-1|HUMAUANTIG|Nog2|Nug2 | ||
| Type: protein-coding gene | Cytoband: 1p34.3 | ||
| Entrez ID: 29889 | HGNC ID: HGNC:29925 | Ensembl Gene: ENSG00000134697 | OMIM ID: 609365 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of GNL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNL2 | 29889 | 201948_at | 0.5479 | 0.2874 | |
| GSE20347 | GNL2 | 29889 | 201948_at | 0.2800 | 0.1058 | |
| GSE23400 | GNL2 | 29889 | 201948_at | 0.4777 | 0.0000 | |
| GSE26886 | GNL2 | 29889 | 201948_at | 1.0017 | 0.0002 | |
| GSE29001 | GNL2 | 29889 | 201948_at | 0.2801 | 0.3409 | |
| GSE38129 | GNL2 | 29889 | 201948_at | 0.4264 | 0.0019 | |
| GSE45670 | GNL2 | 29889 | 201948_at | 0.2397 | 0.1181 | |
| GSE53622 | GNL2 | 29889 | 49438 | 0.4975 | 0.0000 | |
| GSE53624 | GNL2 | 29889 | 49438 | 0.5450 | 0.0000 | |
| GSE63941 | GNL2 | 29889 | 201948_at | -0.0035 | 0.9948 | |
| GSE77861 | GNL2 | 29889 | 201948_at | 0.3376 | 0.3132 | |
| GSE97050 | GNL2 | 29889 | A_23_P34578 | 0.7492 | 0.1782 | |
| SRP007169 | GNL2 | 29889 | RNAseq | 0.3957 | 0.3579 | |
| SRP008496 | GNL2 | 29889 | RNAseq | 0.3046 | 0.2277 | |
| SRP064894 | GNL2 | 29889 | RNAseq | 0.4423 | 0.0048 | |
| SRP133303 | GNL2 | 29889 | RNAseq | 0.5287 | 0.0039 | |
| SRP159526 | GNL2 | 29889 | RNAseq | 0.2315 | 0.4134 | |
| SRP193095 | GNL2 | 29889 | RNAseq | 0.1980 | 0.0346 | |
| SRP219564 | GNL2 | 29889 | RNAseq | 0.5781 | 0.2498 | |
| TCGA | GNL2 | 29889 | RNAseq | 0.1869 | 0.0003 |
Upregulated datasets: 1; Downregulated datasets: 0.
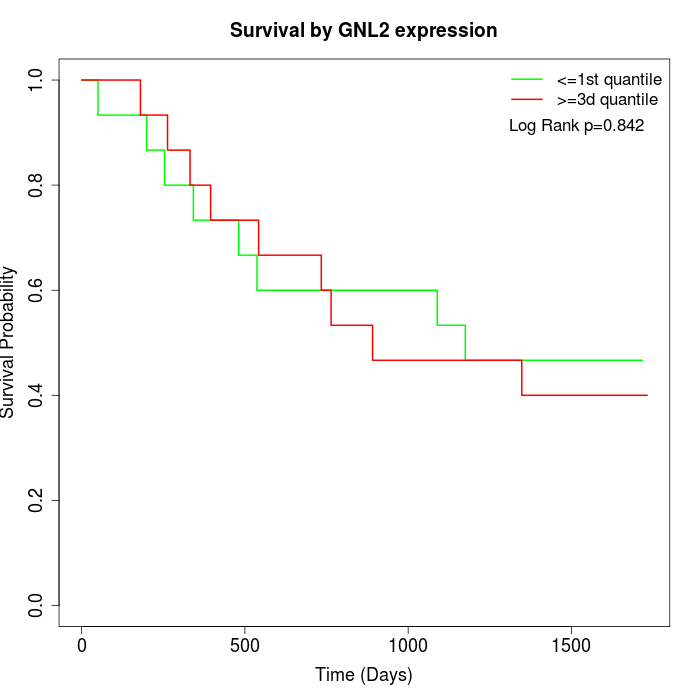
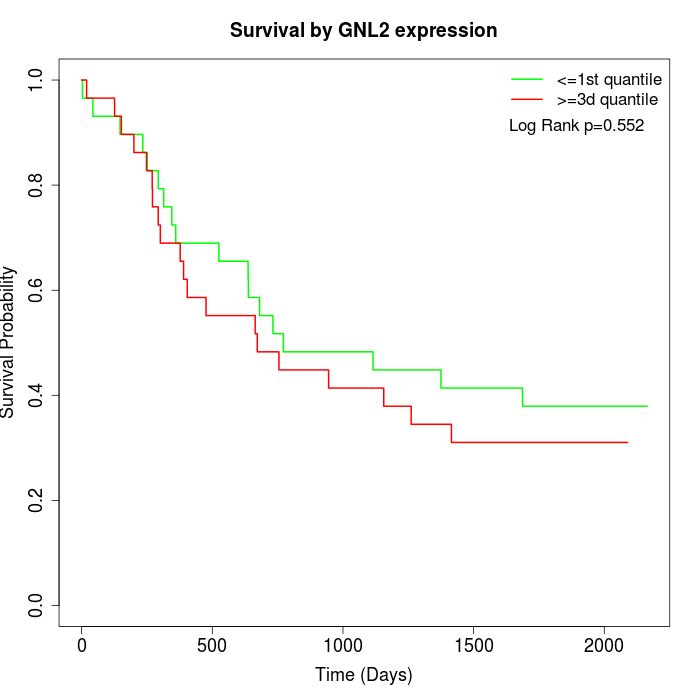
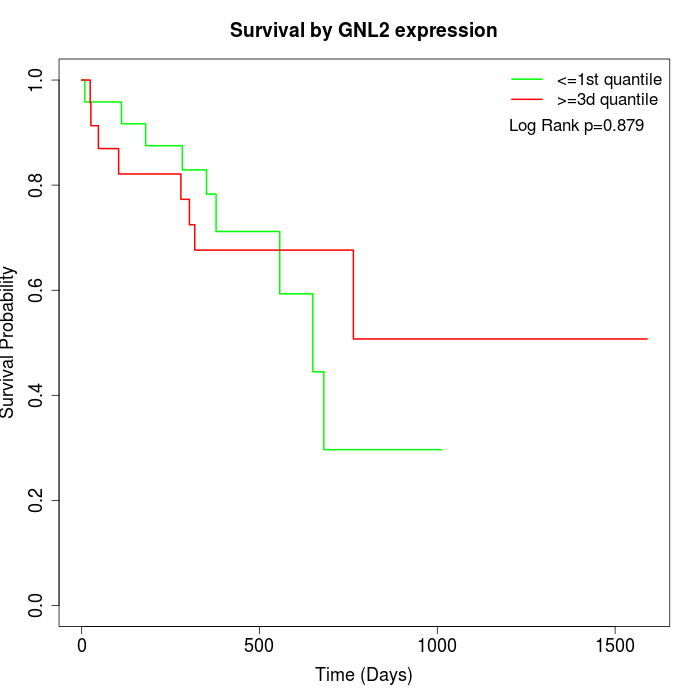
Survival by GNL2 expression:
Note: Click image to view full size file.
Copy number change of GNL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNL2 | 29889 | 2 | 4 | 24 | |
| GSE20123 | GNL2 | 29889 | 2 | 3 | 25 | |
| GSE43470 | GNL2 | 29889 | 7 | 3 | 33 | |
| GSE46452 | GNL2 | 29889 | 4 | 1 | 54 | |
| GSE47630 | GNL2 | 29889 | 8 | 3 | 29 | |
| GSE54993 | GNL2 | 29889 | 1 | 1 | 68 | |
| GSE54994 | GNL2 | 29889 | 11 | 2 | 40 | |
| GSE60625 | GNL2 | 29889 | 0 | 0 | 11 | |
| GSE74703 | GNL2 | 29889 | 6 | 2 | 28 | |
| GSE74704 | GNL2 | 29889 | 1 | 0 | 19 | |
| TCGA | GNL2 | 29889 | 12 | 16 | 68 |
Total number of gains: 54; Total number of losses: 35; Total Number of normals: 399.
Somatic mutations of GNL2:
Generating mutation plots.
Highly correlated genes for GNL2:
Showing top 20/1227 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNL2 | MTHFD1L | 0.763528 | 3 | 0 | 3 |
| GNL2 | ROMO1 | 0.760283 | 3 | 0 | 3 |
| GNL2 | TIGD1 | 0.758437 | 3 | 0 | 3 |
| GNL2 | GPR153 | 0.734973 | 4 | 0 | 4 |
| GNL2 | UCK2 | 0.733825 | 3 | 0 | 3 |
| GNL2 | ALKBH6 | 0.732855 | 3 | 0 | 3 |
| GNL2 | FAM89A | 0.727951 | 3 | 0 | 3 |
| GNL2 | ZFYVE27 | 0.724879 | 3 | 0 | 3 |
| GNL2 | PET117 | 0.723621 | 3 | 0 | 3 |
| GNL2 | RGS1 | 0.711736 | 3 | 0 | 3 |
| GNL2 | MMS19 | 0.697496 | 3 | 0 | 3 |
| GNL2 | UBE2W | 0.697428 | 3 | 0 | 3 |
| GNL2 | ZNF432 | 0.695306 | 3 | 0 | 3 |
| GNL2 | SRSF2 | 0.693519 | 3 | 0 | 3 |
| GNL2 | RAD18 | 0.69237 | 3 | 0 | 3 |
| GNL2 | MED8 | 0.688405 | 3 | 0 | 3 |
| GNL2 | TMEM237 | 0.688117 | 4 | 0 | 3 |
| GNL2 | ANKRD17 | 0.686163 | 4 | 0 | 3 |
| GNL2 | NOP14 | 0.684333 | 7 | 0 | 7 |
| GNL2 | COX17 | 0.682573 | 3 | 0 | 3 |
For details and further investigation, click here