| Full name: glycine N-methyltransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p21.1 | ||
| Entrez ID: 27232 | HGNC ID: HGNC:4415 | Ensembl Gene: ENSG00000124713 | OMIM ID: 606628 |
| Drug and gene relationship at DGIdb | |||
Expression of GNMT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNMT | 27232 | 210328_at | -0.1625 | 0.6867 | |
| GSE20347 | GNMT | 27232 | 210328_at | -0.0843 | 0.3328 | |
| GSE23400 | GNMT | 27232 | 210328_at | -0.1628 | 0.0010 | |
| GSE26886 | GNMT | 27232 | 210328_at | 0.1374 | 0.3222 | |
| GSE29001 | GNMT | 27232 | 210328_at | -0.0933 | 0.4645 | |
| GSE38129 | GNMT | 27232 | 210328_at | -0.2247 | 0.0398 | |
| GSE45670 | GNMT | 27232 | 210328_at | -0.1752 | 0.2231 | |
| GSE53622 | GNMT | 27232 | 43022 | -0.2470 | 0.0528 | |
| GSE53624 | GNMT | 27232 | 43022 | -0.2160 | 0.0615 | |
| GSE63941 | GNMT | 27232 | 210328_at | 0.1527 | 0.5822 | |
| GSE77861 | GNMT | 27232 | 210328_at | -0.0023 | 0.9880 | |
| GSE97050 | GNMT | 27232 | A_23_P7957 | 0.1655 | 0.4168 | |
| TCGA | GNMT | 27232 | RNAseq | -2.3962 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
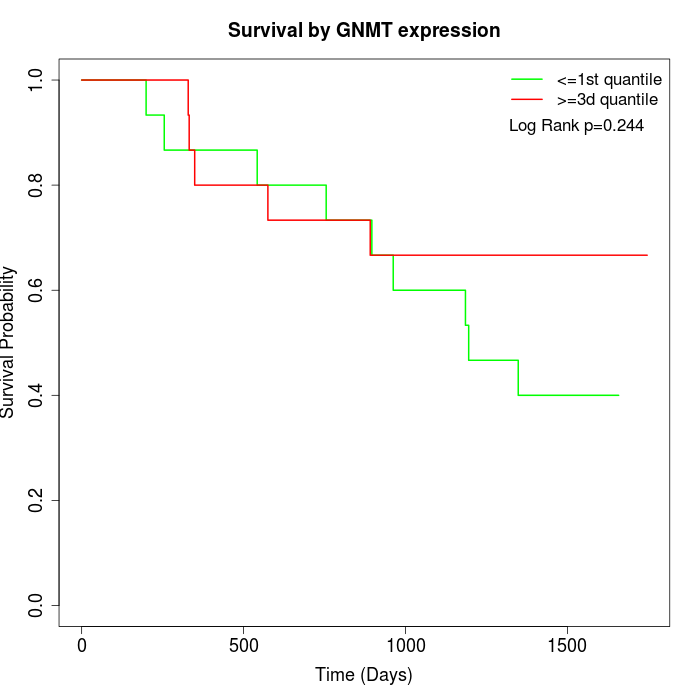
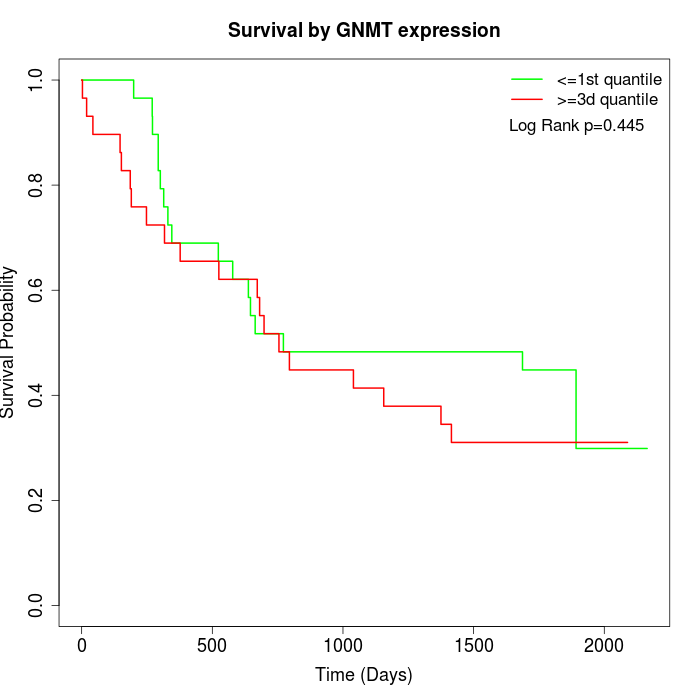
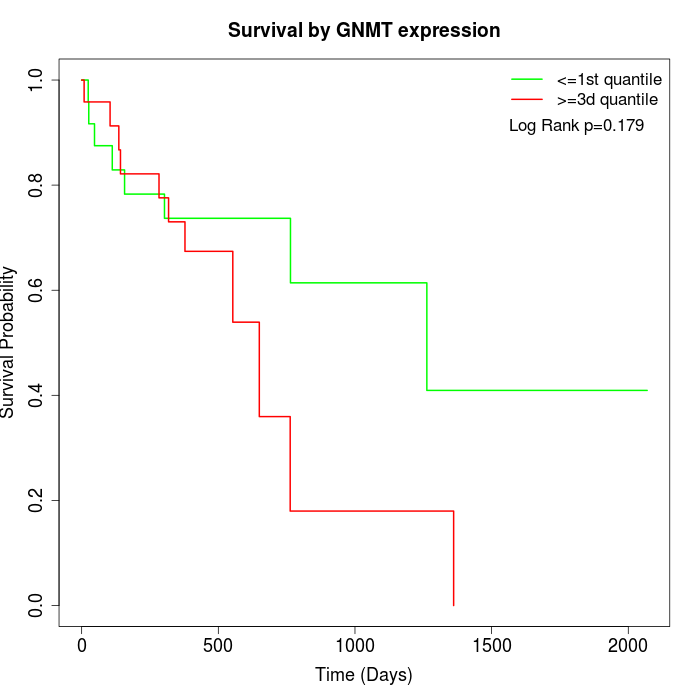
Survival by GNMT expression:
Note: Click image to view full size file.
Copy number change of GNMT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNMT | 27232 | 7 | 0 | 23 | |
| GSE20123 | GNMT | 27232 | 7 | 0 | 23 | |
| GSE43470 | GNMT | 27232 | 6 | 0 | 37 | |
| GSE46452 | GNMT | 27232 | 2 | 9 | 48 | |
| GSE47630 | GNMT | 27232 | 8 | 4 | 28 | |
| GSE54993 | GNMT | 27232 | 3 | 2 | 65 | |
| GSE54994 | GNMT | 27232 | 11 | 6 | 36 | |
| GSE60625 | GNMT | 27232 | 0 | 1 | 10 | |
| GSE74703 | GNMT | 27232 | 6 | 0 | 30 | |
| GSE74704 | GNMT | 27232 | 3 | 0 | 17 | |
| TCGA | GNMT | 27232 | 20 | 13 | 63 |
Total number of gains: 73; Total number of losses: 35; Total Number of normals: 380.
Somatic mutations of GNMT:
Generating mutation plots.
Highly correlated genes for GNMT:
Showing top 20/324 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNMT | PPIL2 | 0.741497 | 3 | 0 | 3 |
| GNMT | FBP2 | 0.722161 | 3 | 0 | 3 |
| GNMT | AQP4 | 0.717052 | 4 | 0 | 4 |
| GNMT | PGAM2 | 0.699847 | 3 | 0 | 3 |
| GNMT | GATA2 | 0.698698 | 4 | 0 | 4 |
| GNMT | MYOZ3 | 0.684631 | 5 | 0 | 5 |
| GNMT | SLC37A1 | 0.683705 | 4 | 0 | 3 |
| GNMT | RBFADN | 0.682141 | 3 | 0 | 3 |
| GNMT | AIRE | 0.68055 | 4 | 0 | 3 |
| GNMT | MUC5AC | 0.680296 | 4 | 0 | 4 |
| GNMT | HECW1 | 0.676495 | 5 | 0 | 4 |
| GNMT | MAPK8IP1 | 0.674185 | 3 | 0 | 3 |
| GNMT | CALCR | 0.670307 | 5 | 0 | 4 |
| GNMT | HAAO | 0.666299 | 5 | 0 | 5 |
| GNMT | GDPD5 | 0.665538 | 4 | 0 | 3 |
| GNMT | UBN2 | 0.657283 | 3 | 0 | 3 |
| GNMT | LCT | 0.657104 | 6 | 0 | 5 |
| GNMT | SULT1B1 | 0.655853 | 4 | 0 | 4 |
| GNMT | ORM1 | 0.655735 | 4 | 0 | 3 |
| GNMT | PPP2R2B | 0.654962 | 4 | 0 | 3 |
For details and further investigation, click here