| Full name: phosphoglycerate mutase 2 | Alias Symbol: PGAM-M | ||
| Type: protein-coding gene | Cytoband: 7p13 | ||
| Entrez ID: 5224 | HGNC ID: HGNC:8889 | Ensembl Gene: ENSG00000164708 | OMIM ID: 612931 |
| Drug and gene relationship at DGIdb | |||
Expression of PGAM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGAM2 | 5224 | 205736_at | 0.0600 | 0.8508 | |
| GSE20347 | PGAM2 | 5224 | 205736_at | -0.0923 | 0.1887 | |
| GSE23400 | PGAM2 | 5224 | 205736_at | -0.1877 | 0.0000 | |
| GSE26886 | PGAM2 | 5224 | 205736_at | -0.0331 | 0.8425 | |
| GSE29001 | PGAM2 | 5224 | 205736_at | -0.0623 | 0.6299 | |
| GSE38129 | PGAM2 | 5224 | 205736_at | -0.2456 | 0.0067 | |
| GSE45670 | PGAM2 | 5224 | 205736_at | -0.1481 | 0.3392 | |
| GSE63941 | PGAM2 | 5224 | 205736_at | 0.2337 | 0.1919 | |
| GSE77861 | PGAM2 | 5224 | 205736_at | -0.0858 | 0.5764 | |
| GSE97050 | PGAM2 | 5224 | A_33_P3268313 | -0.4088 | 0.2079 | |
| TCGA | PGAM2 | 5224 | RNAseq | -0.2538 | 0.1791 |
Upregulated datasets: 0; Downregulated datasets: 0.
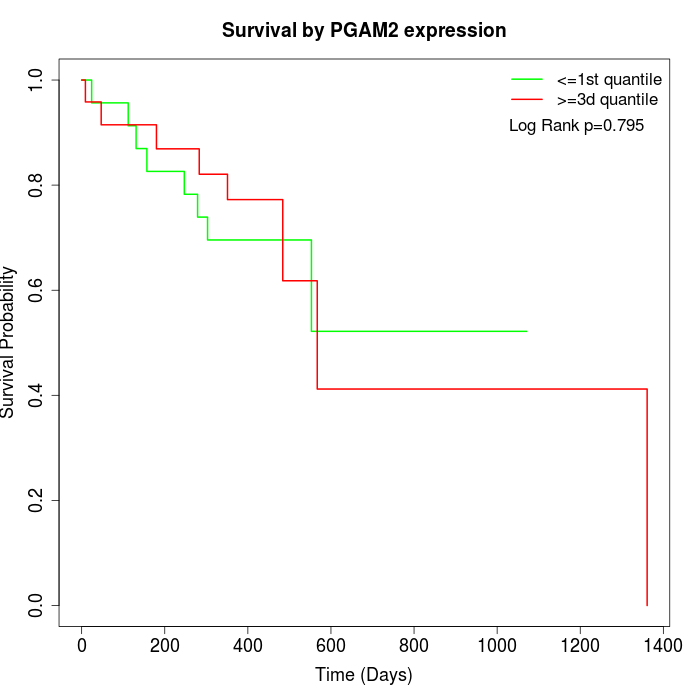
Survival by PGAM2 expression:
Note: Click image to view full size file.
Copy number change of PGAM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGAM2 | 5224 | 14 | 0 | 16 | |
| GSE20123 | PGAM2 | 5224 | 13 | 0 | 17 | |
| GSE43470 | PGAM2 | 5224 | 4 | 0 | 39 | |
| GSE46452 | PGAM2 | 5224 | 11 | 1 | 47 | |
| GSE47630 | PGAM2 | 5224 | 8 | 1 | 31 | |
| GSE54993 | PGAM2 | 5224 | 0 | 8 | 62 | |
| GSE54994 | PGAM2 | 5224 | 17 | 3 | 33 | |
| GSE60625 | PGAM2 | 5224 | 0 | 0 | 11 | |
| GSE74703 | PGAM2 | 5224 | 4 | 0 | 32 | |
| GSE74704 | PGAM2 | 5224 | 7 | 0 | 13 | |
| TCGA | PGAM2 | 5224 | 51 | 7 | 38 |
Total number of gains: 129; Total number of losses: 20; Total Number of normals: 339.
Somatic mutations of PGAM2:
Generating mutation plots.
Highly correlated genes for PGAM2:
Showing top 20/373 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGAM2 | LRFN2 | 0.820084 | 3 | 0 | 3 |
| PGAM2 | FAM181A | 0.732907 | 3 | 0 | 3 |
| PGAM2 | RHPN1 | 0.72706 | 3 | 0 | 3 |
| PGAM2 | PGPEP1 | 0.714344 | 3 | 0 | 3 |
| PGAM2 | RCOR2 | 0.708461 | 4 | 0 | 4 |
| PGAM2 | GNMT | 0.699847 | 3 | 0 | 3 |
| PGAM2 | ARMC5 | 0.690805 | 4 | 0 | 3 |
| PGAM2 | PGC | 0.687541 | 4 | 0 | 4 |
| PGAM2 | UPB1 | 0.686402 | 3 | 0 | 3 |
| PGAM2 | KCNC3 | 0.682394 | 4 | 0 | 3 |
| PGAM2 | MYLK2 | 0.682242 | 4 | 0 | 3 |
| PGAM2 | C20orf141 | 0.680512 | 4 | 0 | 3 |
| PGAM2 | LRRIQ3 | 0.677994 | 3 | 0 | 3 |
| PGAM2 | PPP2R2B | 0.676949 | 3 | 0 | 3 |
| PGAM2 | FFAR1 | 0.675935 | 3 | 0 | 3 |
| PGAM2 | PLA2G4F | 0.675883 | 3 | 0 | 3 |
| PGAM2 | BMP10 | 0.674744 | 4 | 0 | 4 |
| PGAM2 | MT3 | 0.672146 | 3 | 0 | 3 |
| PGAM2 | SHD | 0.666222 | 3 | 0 | 3 |
| PGAM2 | OR3A3 | 0.666008 | 4 | 0 | 4 |
For details and further investigation, click here