| Full name: G protein-coupled bile acid receptor 1 | Alias Symbol: BG37|GPCR|TGR5|M-BAR|GPCR19|GPR131|MGC40597 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 151306 | HGNC ID: HGNC:19680 | Ensembl Gene: ENSG00000179921 | OMIM ID: 610147 |
| Drug and gene relationship at DGIdb | |||
Expression of GPBAR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPBAR1 | 151306 | 1552501_a_at | 0.1301 | 0.5204 | |
| GSE26886 | GPBAR1 | 151306 | 1552501_a_at | -0.1319 | 0.2653 | |
| GSE45670 | GPBAR1 | 151306 | 1552501_a_at | 0.0262 | 0.7452 | |
| GSE53622 | GPBAR1 | 151306 | 147488 | 0.1513 | 0.0362 | |
| GSE53624 | GPBAR1 | 151306 | 147488 | 0.1979 | 0.0018 | |
| GSE63941 | GPBAR1 | 151306 | 1552501_a_at | 0.1800 | 0.1719 | |
| GSE77861 | GPBAR1 | 151306 | 1552501_a_at | -0.0569 | 0.6833 | |
| GSE97050 | GPBAR1 | 151306 | A_23_P400378 | -0.0511 | 0.8714 | |
| SRP159526 | GPBAR1 | 151306 | RNAseq | -0.1321 | 0.8529 | |
| TCGA | GPBAR1 | 151306 | RNAseq | -1.2166 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
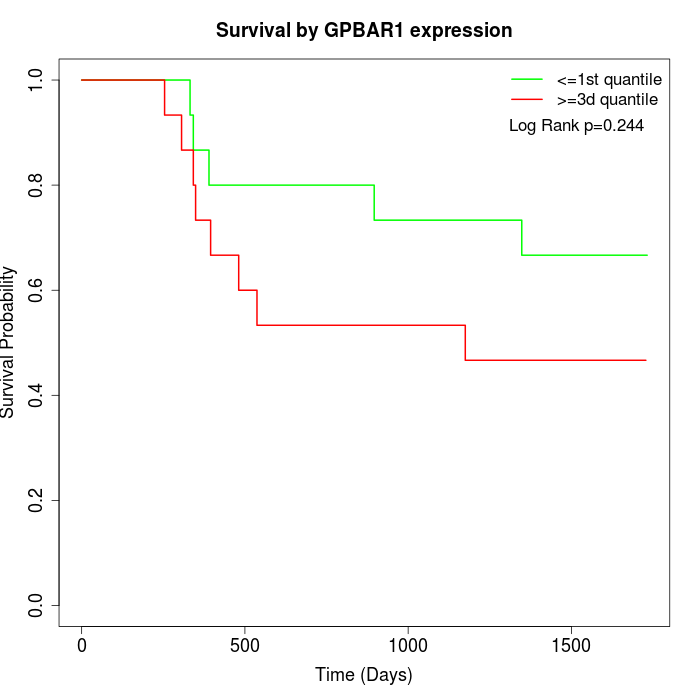
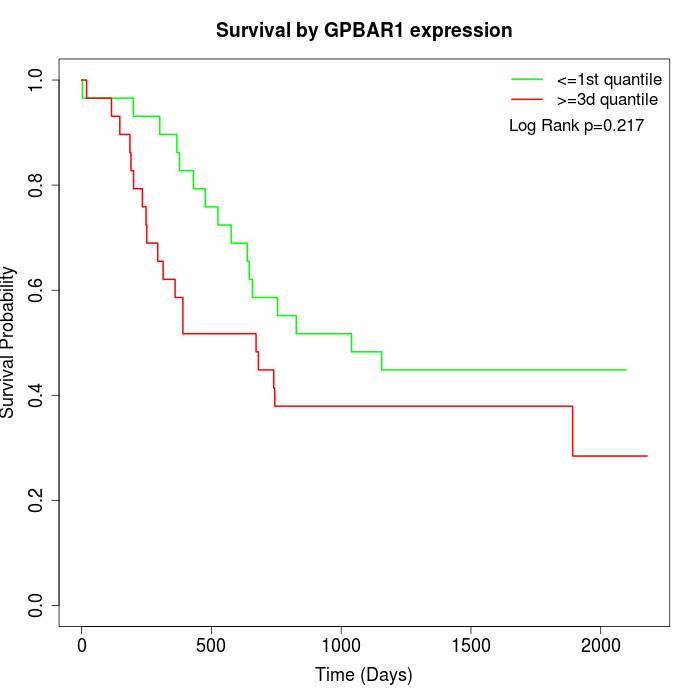
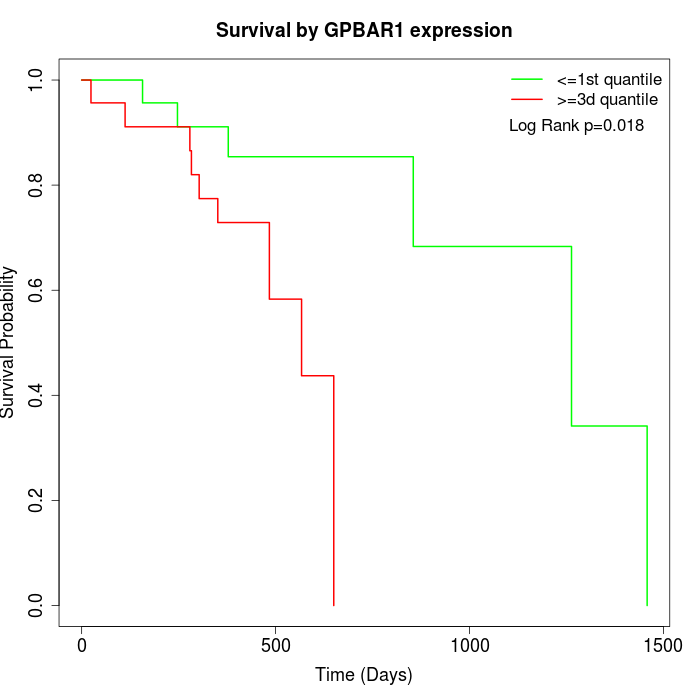
Survival by GPBAR1 expression:
Note: Click image to view full size file.
Copy number change of GPBAR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPBAR1 | 151306 | 1 | 14 | 15 | |
| GSE20123 | GPBAR1 | 151306 | 1 | 13 | 16 | |
| GSE43470 | GPBAR1 | 151306 | 1 | 7 | 35 | |
| GSE46452 | GPBAR1 | 151306 | 0 | 5 | 54 | |
| GSE47630 | GPBAR1 | 151306 | 4 | 5 | 31 | |
| GSE54993 | GPBAR1 | 151306 | 1 | 2 | 67 | |
| GSE54994 | GPBAR1 | 151306 | 8 | 10 | 35 | |
| GSE60625 | GPBAR1 | 151306 | 0 | 3 | 8 | |
| GSE74703 | GPBAR1 | 151306 | 1 | 5 | 30 | |
| GSE74704 | GPBAR1 | 151306 | 1 | 7 | 12 | |
| TCGA | GPBAR1 | 151306 | 11 | 27 | 58 |
Total number of gains: 29; Total number of losses: 98; Total Number of normals: 361.
Somatic mutations of GPBAR1:
Generating mutation plots.
Highly correlated genes for GPBAR1:
Showing top 20/91 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPBAR1 | REG3A | 0.73936 | 3 | 0 | 3 |
| GPBAR1 | HOXD9 | 0.723313 | 3 | 0 | 3 |
| GPBAR1 | ZNF598 | 0.710477 | 3 | 0 | 3 |
| GPBAR1 | LRFN2 | 0.708147 | 3 | 0 | 3 |
| GPBAR1 | SPEF1 | 0.706588 | 3 | 0 | 3 |
| GPBAR1 | MUC8 | 0.703964 | 3 | 0 | 3 |
| GPBAR1 | TMEM52 | 0.701557 | 3 | 0 | 3 |
| GPBAR1 | NOBOX | 0.701237 | 3 | 0 | 3 |
| GPBAR1 | SNAI1 | 0.697417 | 3 | 0 | 3 |
| GPBAR1 | OPTC | 0.696666 | 3 | 0 | 3 |
| GPBAR1 | ANKRD53 | 0.691362 | 4 | 0 | 3 |
| GPBAR1 | SIM1 | 0.680802 | 3 | 0 | 3 |
| GPBAR1 | DBF4B | 0.678298 | 3 | 0 | 3 |
| GPBAR1 | NOC4L | 0.678119 | 3 | 0 | 3 |
| GPBAR1 | PNMA3 | 0.677251 | 3 | 0 | 3 |
| GPBAR1 | TMEM221 | 0.671938 | 3 | 0 | 3 |
| GPBAR1 | SLC22A8 | 0.671458 | 4 | 0 | 3 |
| GPBAR1 | KCNH4 | 0.671227 | 4 | 0 | 4 |
| GPBAR1 | TMEM105 | 0.670103 | 3 | 0 | 3 |
| GPBAR1 | CEP104 | 0.668885 | 3 | 0 | 3 |
For details and further investigation, click here