| Full name: leucine rich repeat and fibronectin type III domain containing 2 | Alias Symbol: FIGLER2 | ||
| Type: protein-coding gene | Cytoband: 6p21.2-p21.1 | ||
| Entrez ID: 57497 | HGNC ID: HGNC:21226 | Ensembl Gene: ENSG00000156564 | OMIM ID: 612808 |
| Drug and gene relationship at DGIdb | |||
Expression of LRFN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LRFN2 | 57497 | 232697_at | -0.1536 | 0.6286 | |
| GSE26886 | LRFN2 | 57497 | 232697_at | -0.0349 | 0.7967 | |
| GSE45670 | LRFN2 | 57497 | 232697_at | -0.0793 | 0.4081 | |
| GSE53622 | LRFN2 | 57497 | 83413 | -0.0105 | 0.9063 | |
| GSE53624 | LRFN2 | 57497 | 83413 | -0.3455 | 0.0012 | |
| GSE63941 | LRFN2 | 57497 | 232697_at | 0.3483 | 0.1143 | |
| GSE77861 | LRFN2 | 57497 | 232697_at | -0.1188 | 0.4287 | |
| GSE97050 | LRFN2 | 57497 | A_32_P82111 | -0.0069 | 0.9806 | |
| SRP159526 | LRFN2 | 57497 | RNAseq | -3.8295 | 0.0000 | |
| TCGA | LRFN2 | 57497 | RNAseq | -0.6319 | 0.3375 |
Upregulated datasets: 0; Downregulated datasets: 1.
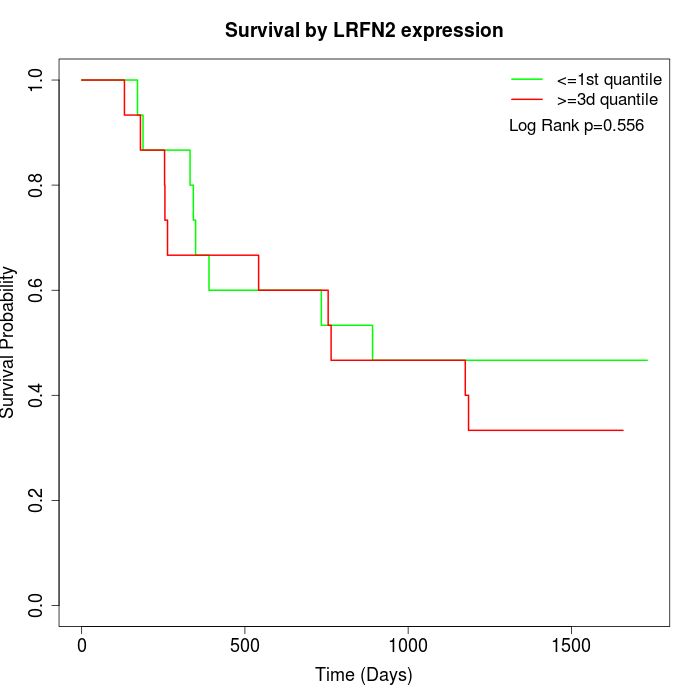
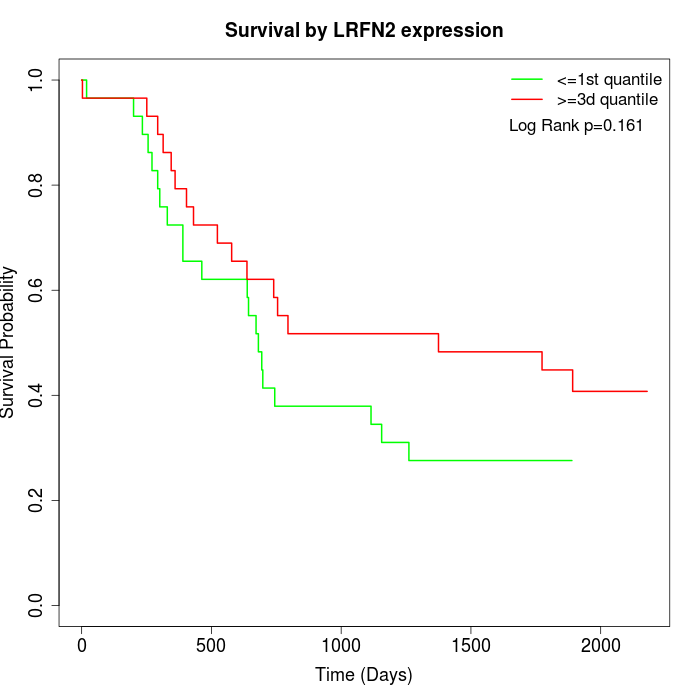
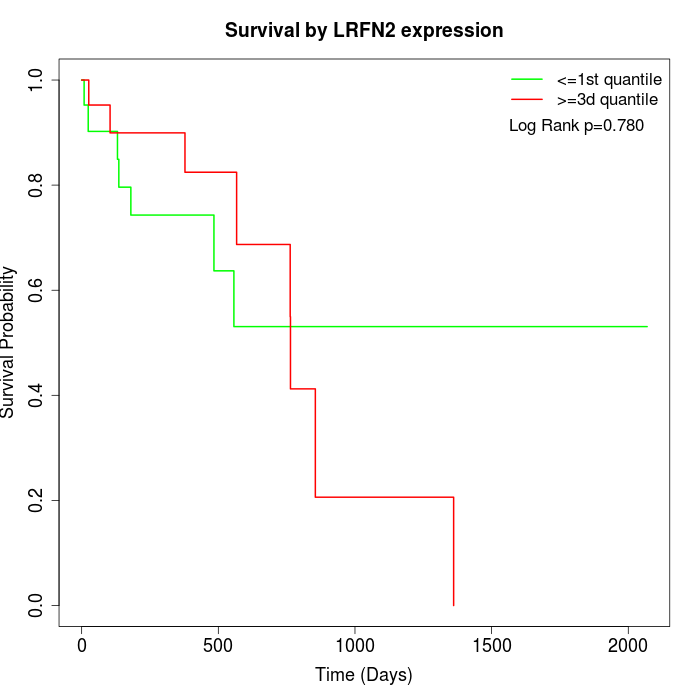
Survival by LRFN2 expression:
Note: Click image to view full size file.
Copy number change of LRFN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LRFN2 | 57497 | 3 | 3 | 24 | |
| GSE20123 | LRFN2 | 57497 | 3 | 3 | 24 | |
| GSE43470 | LRFN2 | 57497 | 6 | 0 | 37 | |
| GSE46452 | LRFN2 | 57497 | 2 | 9 | 48 | |
| GSE47630 | LRFN2 | 57497 | 8 | 4 | 28 | |
| GSE54993 | LRFN2 | 57497 | 3 | 2 | 65 | |
| GSE54994 | LRFN2 | 57497 | 10 | 4 | 39 | |
| GSE60625 | LRFN2 | 57497 | 0 | 1 | 10 | |
| GSE74703 | LRFN2 | 57497 | 6 | 0 | 30 | |
| GSE74704 | LRFN2 | 57497 | 1 | 3 | 16 | |
| TCGA | LRFN2 | 57497 | 20 | 14 | 62 |
Total number of gains: 62; Total number of losses: 43; Total Number of normals: 383.
Somatic mutations of LRFN2:
Generating mutation plots.
Highly correlated genes for LRFN2:
Showing top 20/336 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LRFN2 | PGAM2 | 0.820084 | 3 | 0 | 3 |
| LRFN2 | NPEPL1 | 0.797821 | 3 | 0 | 3 |
| LRFN2 | HTR3A | 0.793595 | 3 | 0 | 3 |
| LRFN2 | SLC6A18 | 0.77834 | 3 | 0 | 3 |
| LRFN2 | PCDH1 | 0.772695 | 3 | 0 | 3 |
| LRFN2 | DUOXA2 | 0.771874 | 3 | 0 | 3 |
| LRFN2 | FBXL15 | 0.771672 | 3 | 0 | 3 |
| LRFN2 | KRTAP6-1 | 0.759092 | 3 | 0 | 3 |
| LRFN2 | UCK1 | 0.756588 | 3 | 0 | 3 |
| LRFN2 | PPIL2 | 0.751958 | 3 | 0 | 3 |
| LRFN2 | TMEM63A | 0.750149 | 3 | 0 | 3 |
| LRFN2 | TM6SF2 | 0.749245 | 3 | 0 | 3 |
| LRFN2 | ARMC7 | 0.747957 | 3 | 0 | 3 |
| LRFN2 | STRC | 0.738534 | 3 | 0 | 3 |
| LRFN2 | CELF3 | 0.737529 | 3 | 0 | 3 |
| LRFN2 | LTC4S | 0.736661 | 3 | 0 | 3 |
| LRFN2 | EVX1 | 0.734222 | 3 | 0 | 3 |
| LRFN2 | GRIK4 | 0.728974 | 3 | 0 | 3 |
| LRFN2 | TTLL9 | 0.728882 | 4 | 0 | 4 |
| LRFN2 | ZMYND10 | 0.727761 | 3 | 0 | 3 |
For details and further investigation, click here