| Full name: G protein-coupled receptor 61 | Alias Symbol: BALGR | ||
| Type: protein-coding gene | Cytoband: 1p13.3 | ||
| Entrez ID: 83873 | HGNC ID: HGNC:13300 | Ensembl Gene: ENSG00000156097 | OMIM ID: 606916 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of GPR61:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPR61 | 83873 | 224531_at | 0.0226 | 0.9349 | |
| GSE26886 | GPR61 | 83873 | 224531_at | -0.0843 | 0.3953 | |
| GSE45670 | GPR61 | 83873 | 224531_at | 0.0999 | 0.2856 | |
| GSE53622 | GPR61 | 83873 | 33498 | 0.0501 | 0.5722 | |
| GSE53624 | GPR61 | 83873 | 33498 | -0.0200 | 0.8576 | |
| GSE63941 | GPR61 | 83873 | 224531_at | 0.1156 | 0.4920 | |
| GSE77861 | GPR61 | 83873 | 224531_at | -0.0285 | 0.7815 | |
| GSE97050 | GPR61 | 83873 | A_23_P326303 | 0.1283 | 0.5970 | |
| TCGA | GPR61 | 83873 | RNAseq | -0.0834 | 0.8914 |
Upregulated datasets: 0; Downregulated datasets: 0.
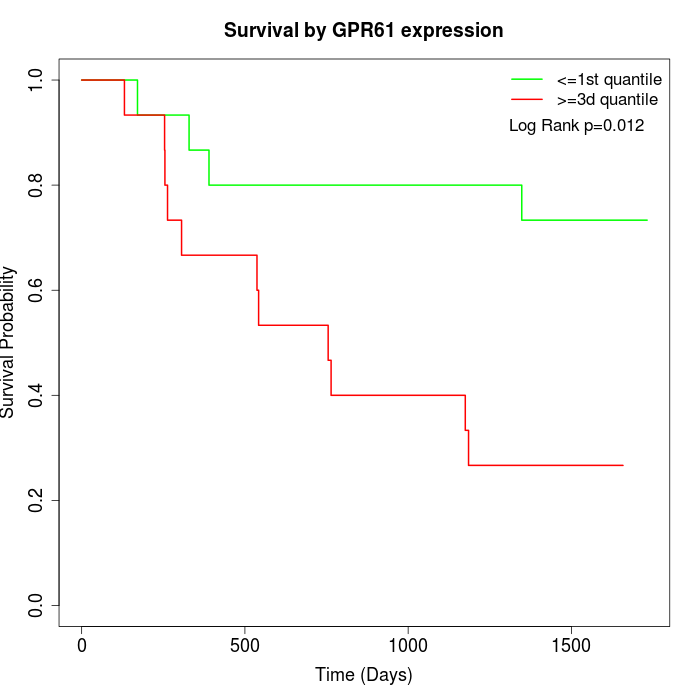
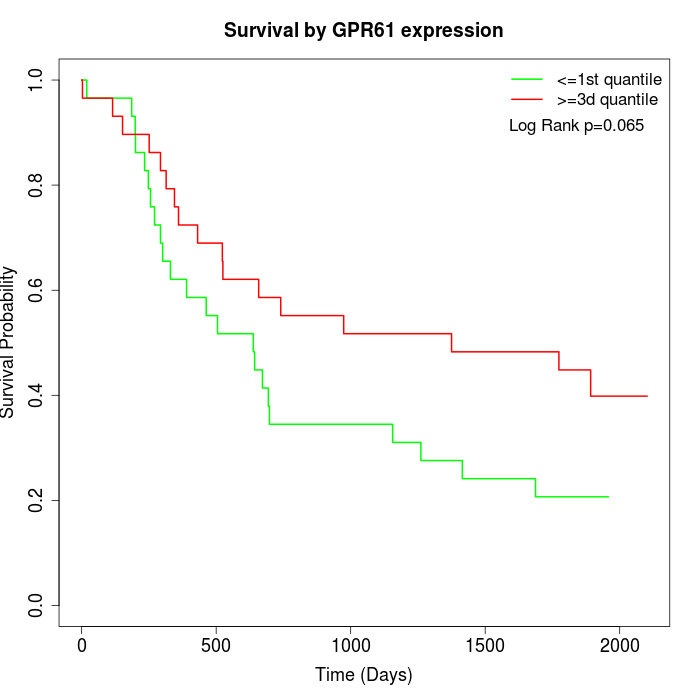
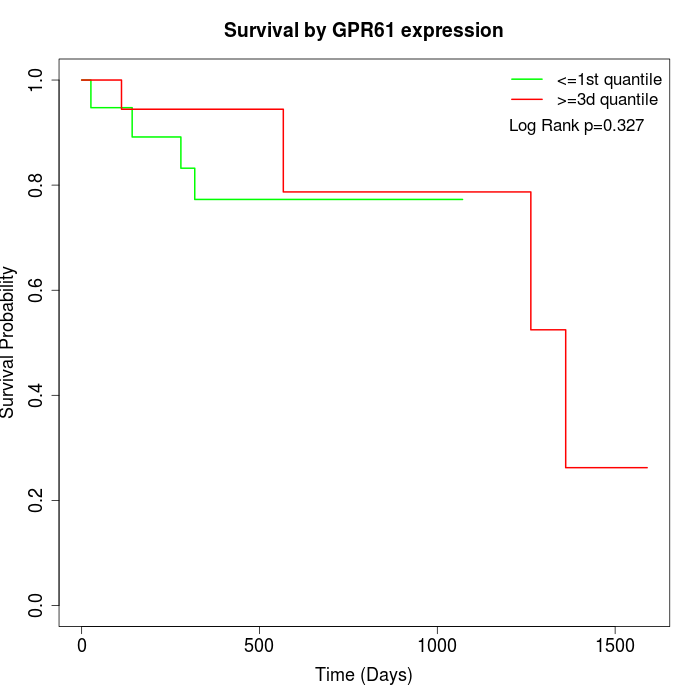
Survival by GPR61 expression:
Note: Click image to view full size file.
Copy number change of GPR61:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPR61 | 83873 | 0 | 12 | 18 | |
| GSE20123 | GPR61 | 83873 | 0 | 11 | 19 | |
| GSE43470 | GPR61 | 83873 | 0 | 9 | 34 | |
| GSE46452 | GPR61 | 83873 | 2 | 1 | 56 | |
| GSE47630 | GPR61 | 83873 | 9 | 5 | 26 | |
| GSE54993 | GPR61 | 83873 | 0 | 1 | 69 | |
| GSE54994 | GPR61 | 83873 | 7 | 4 | 42 | |
| GSE60625 | GPR61 | 83873 | 0 | 0 | 11 | |
| GSE74703 | GPR61 | 83873 | 0 | 7 | 29 | |
| GSE74704 | GPR61 | 83873 | 0 | 7 | 13 | |
| TCGA | GPR61 | 83873 | 8 | 26 | 62 |
Total number of gains: 26; Total number of losses: 83; Total Number of normals: 379.
Somatic mutations of GPR61:
Generating mutation plots.
Highly correlated genes for GPR61:
Showing top 20/129 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPR61 | MYBPC3 | 0.792797 | 3 | 0 | 3 |
| GPR61 | CD300LG | 0.751386 | 3 | 0 | 3 |
| GPR61 | RASL10B | 0.747756 | 3 | 0 | 3 |
| GPR61 | CLDN9 | 0.741923 | 3 | 0 | 3 |
| GPR61 | PNMA3 | 0.72873 | 3 | 0 | 3 |
| GPR61 | NEUROG1 | 0.71452 | 3 | 0 | 3 |
| GPR61 | SPACA3 | 0.711346 | 3 | 0 | 3 |
| GPR61 | KIRREL2 | 0.702875 | 3 | 0 | 3 |
| GPR61 | KCNQ4 | 0.70182 | 3 | 0 | 3 |
| GPR61 | TMEM105 | 0.699907 | 3 | 0 | 3 |
| GPR61 | FFAR1 | 0.698304 | 3 | 0 | 3 |
| GPR61 | ZBED3-AS1 | 0.6834 | 3 | 0 | 3 |
| GPR61 | KCNK3 | 0.681135 | 3 | 0 | 3 |
| GPR61 | CHMP6 | 0.676419 | 3 | 0 | 3 |
| GPR61 | CHD5 | 0.675429 | 3 | 0 | 3 |
| GPR61 | CPLX2 | 0.671835 | 3 | 0 | 3 |
| GPR61 | MYOD1 | 0.670339 | 4 | 0 | 3 |
| GPR61 | RIMS4 | 0.667532 | 3 | 0 | 3 |
| GPR61 | CADPS | 0.665729 | 4 | 0 | 4 |
| GPR61 | KCNC3 | 0.664421 | 3 | 0 | 3 |
For details and further investigation, click here