| Full name: GTF2I repeat domain containing 1 | Alias Symbol: MusTRD1|RBAP2|GTF3|WBSCR12|BEN|Cream1 | ||
| Type: protein-coding gene | Cytoband: 7q11.23 | ||
| Entrez ID: 9569 | HGNC ID: HGNC:4661 | Ensembl Gene: ENSG00000006704 | OMIM ID: 604318 |
| Drug and gene relationship at DGIdb | |||
GTF2IRD1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway |
Expression of GTF2IRD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GTF2IRD1 | 9569 | 218412_s_at | 0.0673 | 0.8972 | |
| GSE20347 | GTF2IRD1 | 9569 | 218412_s_at | 0.1030 | 0.4327 | |
| GSE23400 | GTF2IRD1 | 9569 | 218412_s_at | 0.0485 | 0.5735 | |
| GSE26886 | GTF2IRD1 | 9569 | 218412_s_at | 0.5763 | 0.0140 | |
| GSE29001 | GTF2IRD1 | 9569 | 218412_s_at | -0.0248 | 0.9122 | |
| GSE38129 | GTF2IRD1 | 9569 | 218412_s_at | 0.2801 | 0.0601 | |
| GSE45670 | GTF2IRD1 | 9569 | 218412_s_at | 0.1160 | 0.4073 | |
| GSE53622 | GTF2IRD1 | 9569 | 15700 | 0.1713 | 0.0840 | |
| GSE53624 | GTF2IRD1 | 9569 | 15700 | 0.0424 | 0.5224 | |
| GSE63941 | GTF2IRD1 | 9569 | 218412_s_at | 0.2662 | 0.3382 | |
| GSE77861 | GTF2IRD1 | 9569 | 218412_s_at | 0.0528 | 0.8712 | |
| GSE97050 | GTF2IRD1 | 9569 | A_23_P111621 | 0.2031 | 0.5259 | |
| SRP007169 | GTF2IRD1 | 9569 | RNAseq | -0.7953 | 0.0178 | |
| SRP008496 | GTF2IRD1 | 9569 | RNAseq | -0.6772 | 0.0112 | |
| SRP064894 | GTF2IRD1 | 9569 | RNAseq | -0.6003 | 0.0013 | |
| SRP133303 | GTF2IRD1 | 9569 | RNAseq | -0.1654 | 0.3403 | |
| SRP159526 | GTF2IRD1 | 9569 | RNAseq | -0.1950 | 0.3883 | |
| SRP193095 | GTF2IRD1 | 9569 | RNAseq | -0.0388 | 0.8246 | |
| SRP219564 | GTF2IRD1 | 9569 | RNAseq | -0.3232 | 0.5256 | |
| TCGA | GTF2IRD1 | 9569 | RNAseq | 0.1883 | 0.0018 |
Upregulated datasets: 0; Downregulated datasets: 0.
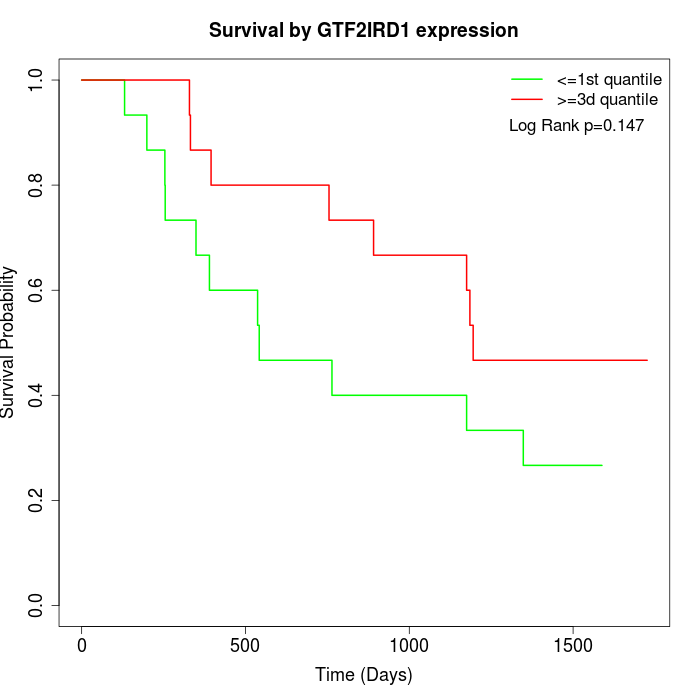
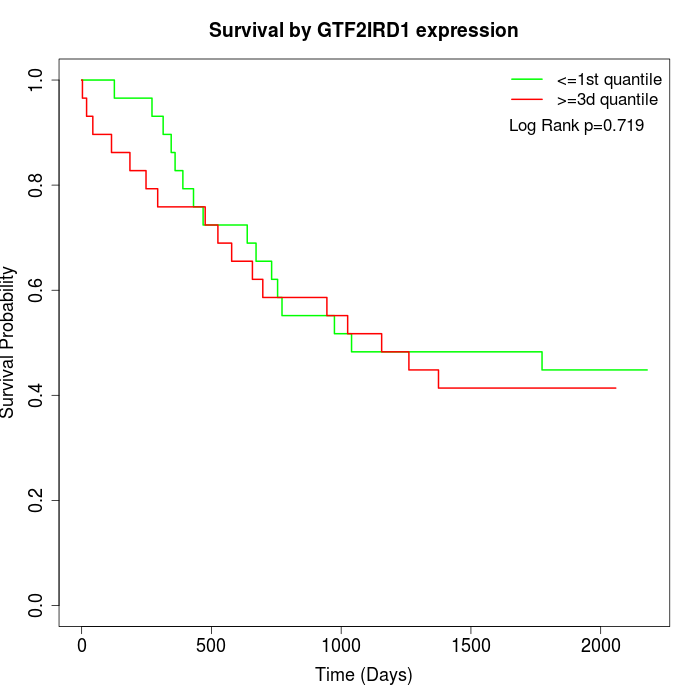
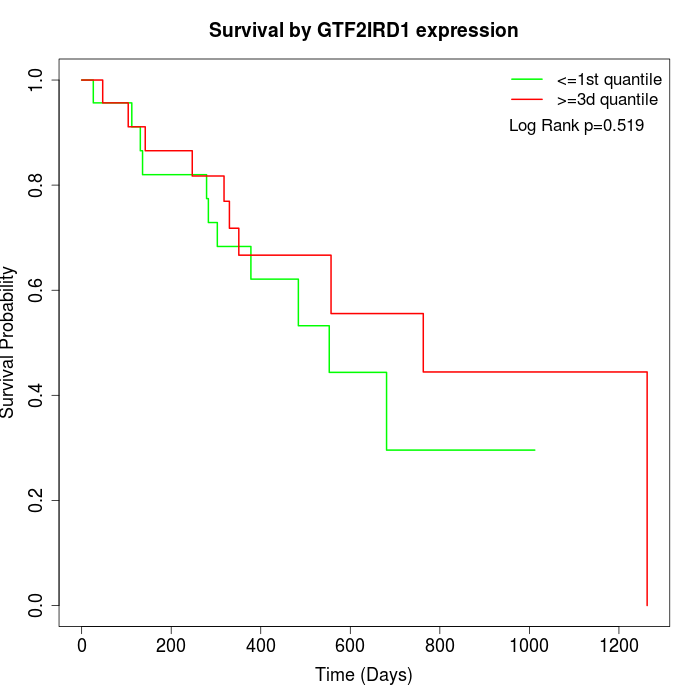
Survival by GTF2IRD1 expression:
Note: Click image to view full size file.
Copy number change of GTF2IRD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GTF2IRD1 | 9569 | 14 | 1 | 15 | |
| GSE20123 | GTF2IRD1 | 9569 | 14 | 1 | 15 | |
| GSE43470 | GTF2IRD1 | 9569 | 3 | 2 | 38 | |
| GSE46452 | GTF2IRD1 | 9569 | 12 | 0 | 47 | |
| GSE47630 | GTF2IRD1 | 9569 | 7 | 2 | 31 | |
| GSE54993 | GTF2IRD1 | 9569 | 2 | 7 | 61 | |
| GSE54994 | GTF2IRD1 | 9569 | 13 | 3 | 37 | |
| GSE60625 | GTF2IRD1 | 9569 | 0 | 0 | 11 | |
| GSE74703 | GTF2IRD1 | 9569 | 3 | 1 | 32 | |
| GSE74704 | GTF2IRD1 | 9569 | 8 | 1 | 11 | |
| TCGA | GTF2IRD1 | 9569 | 45 | 7 | 44 |
Total number of gains: 121; Total number of losses: 25; Total Number of normals: 342.
Somatic mutations of GTF2IRD1:
Generating mutation plots.
Highly correlated genes for GTF2IRD1:
Showing top 20/295 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GTF2IRD1 | ANKRD16 | 0.778927 | 3 | 0 | 3 |
| GTF2IRD1 | TBK1 | 0.765153 | 3 | 0 | 3 |
| GTF2IRD1 | PUSL1 | 0.756464 | 3 | 0 | 3 |
| GTF2IRD1 | DAZAP1 | 0.750176 | 3 | 0 | 3 |
| GTF2IRD1 | CHMP4C | 0.732306 | 3 | 0 | 3 |
| GTF2IRD1 | SMARCD2 | 0.728295 | 4 | 0 | 4 |
| GTF2IRD1 | ANAPC13 | 0.727091 | 3 | 0 | 3 |
| GTF2IRD1 | PSENEN | 0.725602 | 3 | 0 | 3 |
| GTF2IRD1 | CDIPT | 0.715827 | 3 | 0 | 3 |
| GTF2IRD1 | C6orf120 | 0.700015 | 3 | 0 | 3 |
| GTF2IRD1 | DMBT1 | 0.695358 | 3 | 0 | 3 |
| GTF2IRD1 | HOXD8 | 0.693313 | 3 | 0 | 3 |
| GTF2IRD1 | VPS33A | 0.692681 | 3 | 0 | 3 |
| GTF2IRD1 | NUDT19 | 0.68819 | 4 | 0 | 3 |
| GTF2IRD1 | IFT140 | 0.687922 | 3 | 0 | 3 |
| GTF2IRD1 | BAD | 0.687853 | 3 | 0 | 3 |
| GTF2IRD1 | SRP68 | 0.68706 | 4 | 0 | 4 |
| GTF2IRD1 | ZNF184 | 0.684309 | 3 | 0 | 3 |
| GTF2IRD1 | SLC39A11 | 0.683678 | 4 | 0 | 3 |
| GTF2IRD1 | ASH1L | 0.681117 | 3 | 0 | 3 |
For details and further investigation, click here