| Full name: BCL2 associated agonist of cell death | Alias Symbol: BCL2L8|BBC2 | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 572 | HGNC ID: HGNC:936 | Ensembl Gene: ENSG00000002330 | OMIM ID: 603167 |
| Drug and gene relationship at DGIdb | |||
BAD involved pathways:
Expression of BAD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BAD | 572 | 209364_at | 0.1260 | 0.8112 | |
| GSE20347 | BAD | 572 | 1861_at | -0.1085 | 0.6172 | |
| GSE23400 | BAD | 572 | 209364_at | -0.0563 | 0.3975 | |
| GSE26886 | BAD | 572 | 209364_at | 0.9004 | 0.0000 | |
| GSE29001 | BAD | 572 | 209364_at | -0.1490 | 0.5024 | |
| GSE38129 | BAD | 572 | 1861_at | -0.0498 | 0.8182 | |
| GSE45670 | BAD | 572 | 209364_at | -0.0855 | 0.4660 | |
| GSE53622 | BAD | 572 | 41913 | -0.1201 | 0.2264 | |
| GSE53624 | BAD | 572 | 41913 | 0.1618 | 0.0795 | |
| GSE63941 | BAD | 572 | 209364_at | 0.2249 | 0.6286 | |
| GSE77861 | BAD | 572 | 209364_at | 0.1068 | 0.5947 | |
| GSE97050 | BAD | 572 | A_33_P3402076 | -0.0212 | 0.9523 | |
| SRP007169 | BAD | 572 | RNAseq | -1.2454 | 0.0067 | |
| SRP008496 | BAD | 572 | RNAseq | -0.2563 | 0.6136 | |
| SRP064894 | BAD | 572 | RNAseq | -0.3012 | 0.4110 | |
| SRP133303 | BAD | 572 | RNAseq | 0.0826 | 0.7082 | |
| SRP159526 | BAD | 572 | RNAseq | 0.1680 | 0.5393 | |
| SRP193095 | BAD | 572 | RNAseq | -0.3106 | 0.0521 | |
| SRP219564 | BAD | 572 | RNAseq | -0.3952 | 0.3819 | |
| TCGA | BAD | 572 | RNAseq | -0.0884 | 0.1157 |
Upregulated datasets: 0; Downregulated datasets: 1.
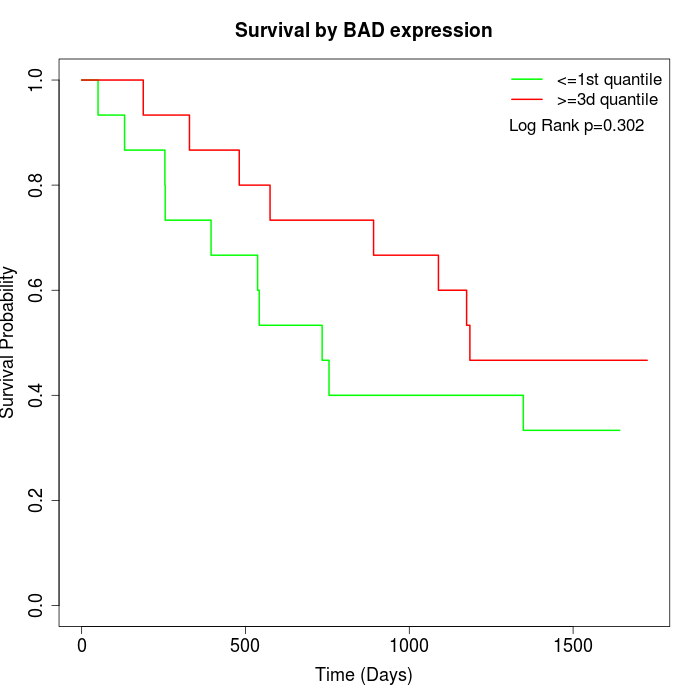
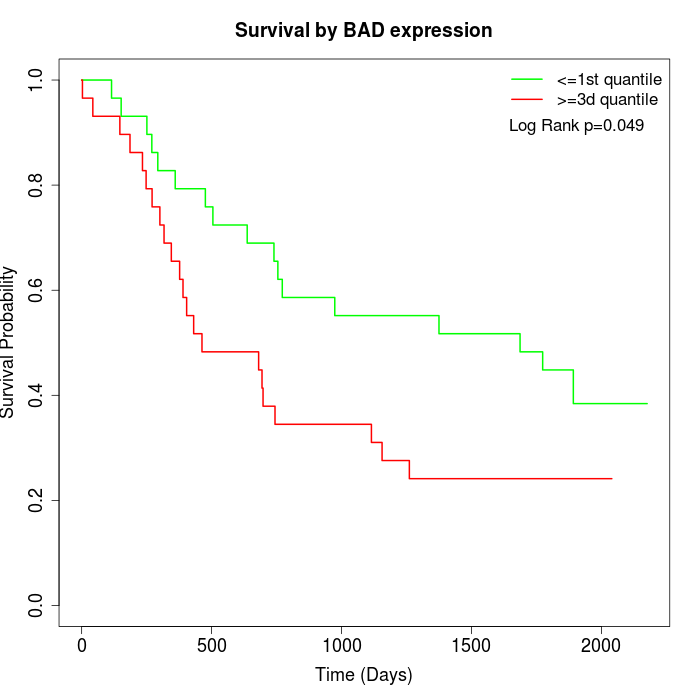
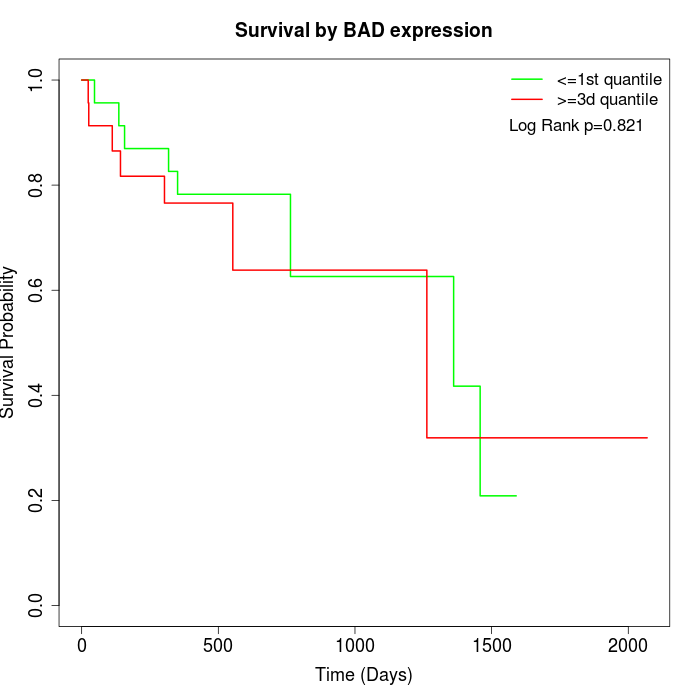
Survival by BAD expression:
Note: Click image to view full size file.
Copy number change of BAD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BAD | 572 | 6 | 5 | 19 | |
| GSE20123 | BAD | 572 | 6 | 5 | 19 | |
| GSE43470 | BAD | 572 | 1 | 3 | 39 | |
| GSE46452 | BAD | 572 | 8 | 4 | 47 | |
| GSE47630 | BAD | 572 | 4 | 6 | 30 | |
| GSE54993 | BAD | 572 | 3 | 0 | 67 | |
| GSE54994 | BAD | 572 | 5 | 5 | 43 | |
| GSE60625 | BAD | 572 | 0 | 4 | 7 | |
| GSE74703 | BAD | 572 | 1 | 1 | 34 | |
| GSE74704 | BAD | 572 | 4 | 3 | 13 | |
| TCGA | BAD | 572 | 20 | 9 | 67 |
Total number of gains: 58; Total number of losses: 45; Total Number of normals: 385.
Somatic mutations of BAD:
Generating mutation plots.
Highly correlated genes for BAD:
Showing top 20/380 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BAD | CPT1A | 0.844211 | 3 | 0 | 3 |
| BAD | DLG5 | 0.774364 | 3 | 0 | 3 |
| BAD | ABCB6 | 0.762586 | 3 | 0 | 3 |
| BAD | HOXA7 | 0.76198 | 3 | 0 | 3 |
| BAD | CPSF6 | 0.75743 | 3 | 0 | 3 |
| BAD | MRPL47 | 0.756284 | 3 | 0 | 3 |
| BAD | FADD | 0.752885 | 3 | 0 | 3 |
| BAD | ITGB8 | 0.750774 | 3 | 0 | 3 |
| BAD | PDPR | 0.747856 | 3 | 0 | 3 |
| BAD | RBM4 | 0.745801 | 3 | 0 | 3 |
| BAD | ZHX2 | 0.733475 | 3 | 0 | 3 |
| BAD | PSMG3 | 0.73284 | 4 | 0 | 4 |
| BAD | ALCAM | 0.73124 | 3 | 0 | 3 |
| BAD | FKTN | 0.73113 | 3 | 0 | 3 |
| BAD | FAM86C1 | 0.727094 | 3 | 0 | 3 |
| BAD | PARL | 0.725471 | 3 | 0 | 3 |
| BAD | LOXL4 | 0.722656 | 3 | 0 | 3 |
| BAD | RNF130 | 0.719562 | 3 | 0 | 3 |
| BAD | MRPL21 | 0.718265 | 3 | 0 | 3 |
| BAD | NCOR2 | 0.717344 | 3 | 0 | 3 |
For details and further investigation, click here