| Full name: melatonin receptor 1B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q14.3 | ||
| Entrez ID: 4544 | HGNC ID: HGNC:7464 | Ensembl Gene: ENSG00000134640 | OMIM ID: 600804 |
| Drug and gene relationship at DGIdb | |||
MTNR1B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04713 | Circadian entrainment |
Expression of MTNR1B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MTNR1B | 4544 | 208516_at | -0.1215 | 0.5600 | |
| GSE20347 | MTNR1B | 4544 | 208516_at | -0.0348 | 0.6979 | |
| GSE23400 | MTNR1B | 4544 | 208516_at | -0.1140 | 0.0008 | |
| GSE26886 | MTNR1B | 4544 | 208516_at | 0.1212 | 0.2490 | |
| GSE29001 | MTNR1B | 4544 | 208516_at | -0.0799 | 0.6614 | |
| GSE38129 | MTNR1B | 4544 | 208516_at | -0.1556 | 0.0169 | |
| GSE45670 | MTNR1B | 4544 | 208516_at | -0.0072 | 0.9345 | |
| GSE53622 | MTNR1B | 4544 | 2738 | 0.1144 | 0.1415 | |
| GSE53624 | MTNR1B | 4544 | 2738 | 0.1389 | 0.0730 | |
| GSE63941 | MTNR1B | 4544 | 208516_at | 0.0827 | 0.5748 | |
| GSE77861 | MTNR1B | 4544 | 208516_at | -0.0135 | 0.9219 | |
| TCGA | MTNR1B | 4544 | RNAseq | 1.2468 | 0.5019 |
Upregulated datasets: 0; Downregulated datasets: 0.
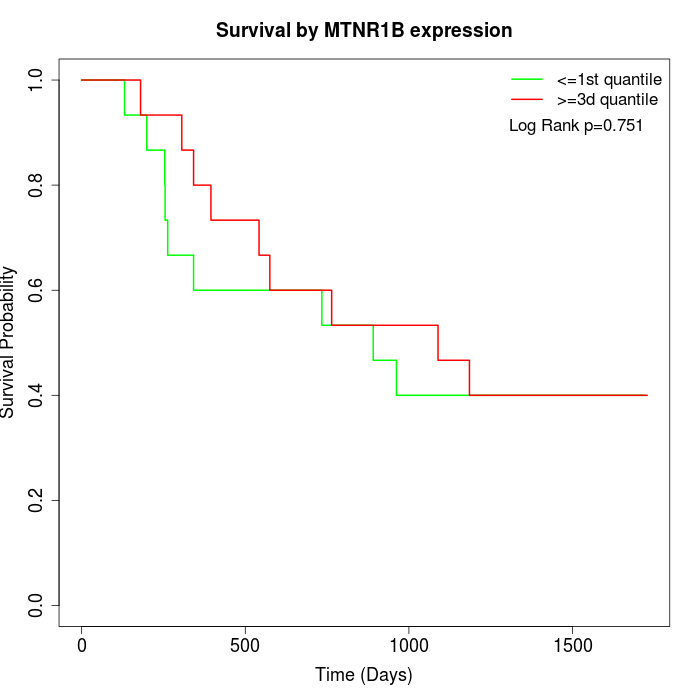
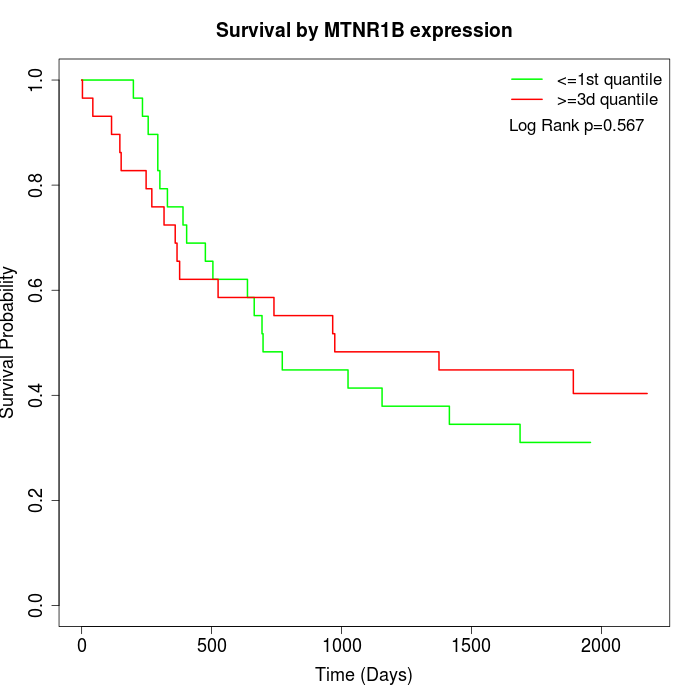
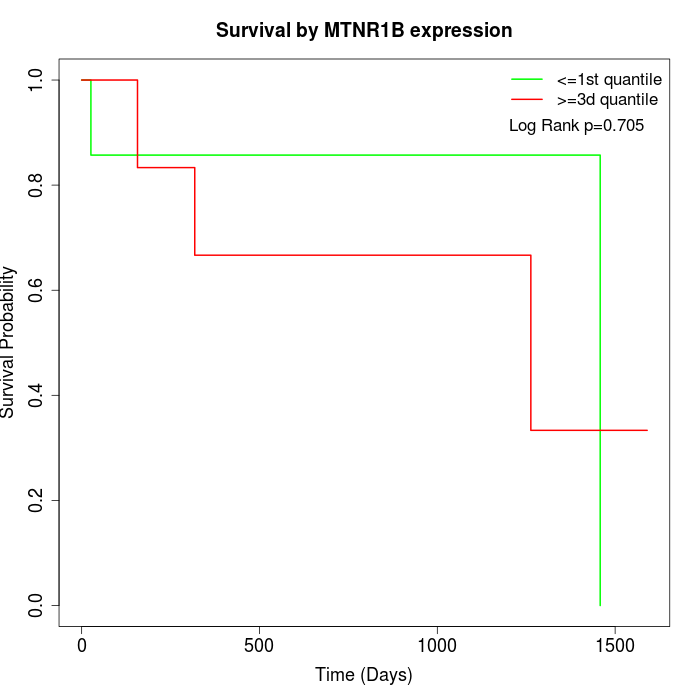
Survival by MTNR1B expression:
Note: Click image to view full size file.
Copy number change of MTNR1B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MTNR1B | 4544 | 3 | 10 | 17 | |
| GSE20123 | MTNR1B | 4544 | 3 | 10 | 17 | |
| GSE43470 | MTNR1B | 4544 | 2 | 5 | 36 | |
| GSE46452 | MTNR1B | 4544 | 5 | 24 | 30 | |
| GSE47630 | MTNR1B | 4544 | 2 | 19 | 19 | |
| GSE54993 | MTNR1B | 4544 | 8 | 2 | 60 | |
| GSE54994 | MTNR1B | 4544 | 5 | 19 | 29 | |
| GSE60625 | MTNR1B | 4544 | 0 | 3 | 8 | |
| GSE74703 | MTNR1B | 4544 | 1 | 4 | 31 | |
| GSE74704 | MTNR1B | 4544 | 3 | 6 | 11 | |
| TCGA | MTNR1B | 4544 | 13 | 40 | 43 |
Total number of gains: 45; Total number of losses: 142; Total Number of normals: 301.
Somatic mutations of MTNR1B:
Generating mutation plots.
Highly correlated genes for MTNR1B:
Showing top 20/655 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MTNR1B | ZMAT4 | 0.759328 | 3 | 0 | 3 |
| MTNR1B | OR10H3 | 0.716425 | 5 | 0 | 5 |
| MTNR1B | CITED1 | 0.700662 | 3 | 0 | 3 |
| MTNR1B | GYPA | 0.698978 | 4 | 0 | 4 |
| MTNR1B | HLA-DOA | 0.697531 | 4 | 0 | 4 |
| MTNR1B | RASL10A | 0.691725 | 3 | 0 | 3 |
| MTNR1B | RASA3 | 0.691608 | 3 | 0 | 3 |
| MTNR1B | CXCR5 | 0.683696 | 4 | 0 | 3 |
| MTNR1B | RUNX2 | 0.678972 | 4 | 0 | 4 |
| MTNR1B | NCR1 | 0.672773 | 5 | 0 | 4 |
| MTNR1B | RORC | 0.669248 | 6 | 0 | 6 |
| MTNR1B | CACNA1S | 0.666218 | 3 | 0 | 3 |
| MTNR1B | GJA4 | 0.665866 | 3 | 0 | 3 |
| MTNR1B | OR12D2 | 0.661618 | 4 | 0 | 4 |
| MTNR1B | LMAN1L | 0.661559 | 4 | 0 | 4 |
| MTNR1B | HTR4 | 0.661487 | 4 | 0 | 4 |
| MTNR1B | PHLDB1 | 0.660216 | 5 | 0 | 4 |
| MTNR1B | TLR4 | 0.658625 | 3 | 0 | 3 |
| MTNR1B | RIC3 | 0.65711 | 4 | 0 | 4 |
| MTNR1B | CGA | 0.653231 | 4 | 0 | 3 |
For details and further investigation, click here