| Full name: hydroxyacyl-CoA dehydrogenase | Alias Symbol: HADH1|SCHAD | ||
| Type: protein-coding gene | Cytoband: 4q25 | ||
| Entrez ID: 3033 | HGNC ID: HGNC:4799 | Ensembl Gene: ENSG00000138796 | OMIM ID: 601609 |
| Drug and gene relationship at DGIdb | |||
Expression of HADH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HADH | 3033 | 201036_s_at | 0.2885 | 0.6418 | |
| GSE20347 | HADH | 3033 | 201036_s_at | -0.1067 | 0.5716 | |
| GSE23400 | HADH | 3033 | 201035_s_at | -0.1107 | 0.0235 | |
| GSE26886 | HADH | 3033 | 201036_s_at | -0.3055 | 0.2503 | |
| GSE29001 | HADH | 3033 | 201036_s_at | -0.0249 | 0.9506 | |
| GSE38129 | HADH | 3033 | 201036_s_at | -0.3960 | 0.0202 | |
| GSE45670 | HADH | 3033 | 201036_s_at | -0.4140 | 0.0350 | |
| GSE53622 | HADH | 3033 | 43192 | -0.7854 | 0.0000 | |
| GSE53624 | HADH | 3033 | 43192 | -0.4809 | 0.0000 | |
| GSE63941 | HADH | 3033 | 201036_s_at | 0.1857 | 0.6821 | |
| GSE77861 | HADH | 3033 | 201036_s_at | -0.4346 | 0.1471 | |
| GSE97050 | HADH | 3033 | A_23_P167227 | -0.7910 | 0.1440 | |
| SRP007169 | HADH | 3033 | RNAseq | -0.3818 | 0.3577 | |
| SRP008496 | HADH | 3033 | RNAseq | -0.8299 | 0.0036 | |
| SRP064894 | HADH | 3033 | RNAseq | -0.3455 | 0.0982 | |
| SRP133303 | HADH | 3033 | RNAseq | 0.2766 | 0.3020 | |
| SRP159526 | HADH | 3033 | RNAseq | -0.2091 | 0.4134 | |
| SRP193095 | HADH | 3033 | RNAseq | -0.5127 | 0.0082 | |
| SRP219564 | HADH | 3033 | RNAseq | -0.5541 | 0.2376 | |
| TCGA | HADH | 3033 | RNAseq | -0.3087 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
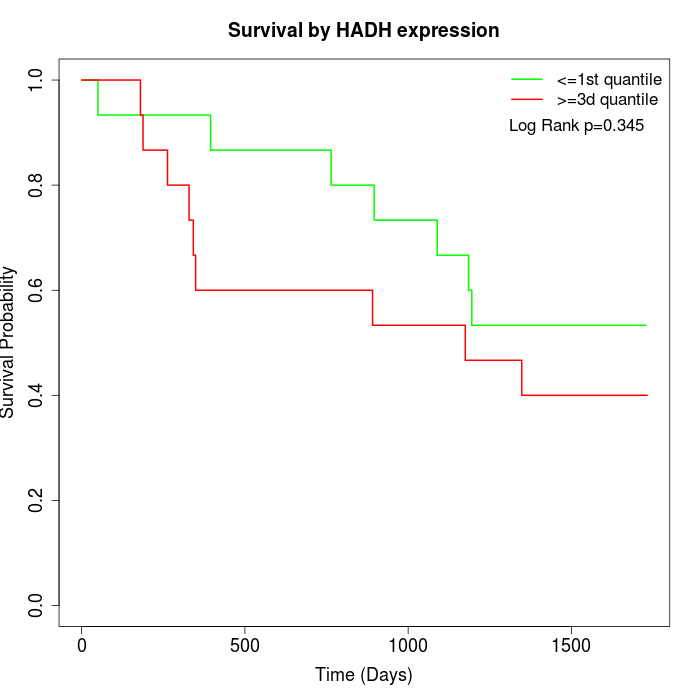
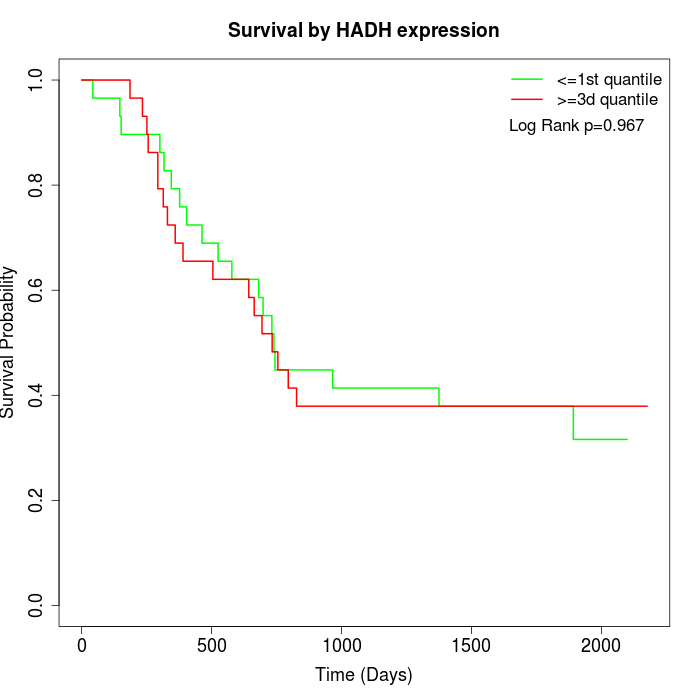
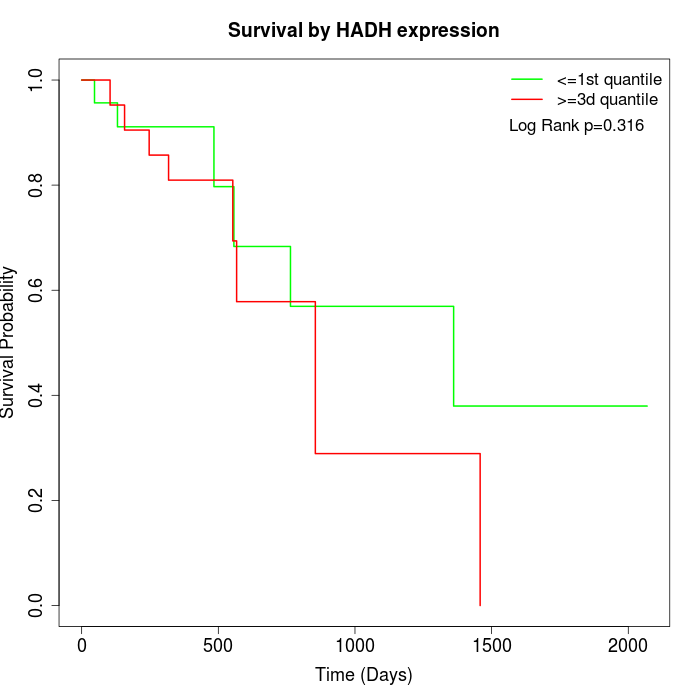
Survival by HADH expression:
Note: Click image to view full size file.
Copy number change of HADH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HADH | 3033 | 0 | 12 | 18 | |
| GSE20123 | HADH | 3033 | 0 | 12 | 18 | |
| GSE43470 | HADH | 3033 | 0 | 13 | 30 | |
| GSE46452 | HADH | 3033 | 1 | 36 | 22 | |
| GSE47630 | HADH | 3033 | 0 | 20 | 20 | |
| GSE54993 | HADH | 3033 | 9 | 0 | 61 | |
| GSE54994 | HADH | 3033 | 1 | 10 | 42 | |
| GSE60625 | HADH | 3033 | 0 | 3 | 8 | |
| GSE74703 | HADH | 3033 | 0 | 11 | 25 | |
| GSE74704 | HADH | 3033 | 0 | 6 | 14 | |
| TCGA | HADH | 3033 | 8 | 35 | 53 |
Total number of gains: 19; Total number of losses: 158; Total Number of normals: 311.
Somatic mutations of HADH:
Generating mutation plots.
Highly correlated genes for HADH:
Showing top 20/679 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HADH | MRPL24 | 0.74911 | 3 | 0 | 3 |
| HADH | ACADSB | 0.720875 | 5 | 0 | 4 |
| HADH | TMEM25 | 0.71802 | 4 | 0 | 3 |
| HADH | ANAPC16 | 0.715076 | 5 | 0 | 4 |
| HADH | RPL14 | 0.709974 | 3 | 0 | 3 |
| HADH | ZNF436 | 0.707524 | 3 | 0 | 3 |
| HADH | UBXN10 | 0.703727 | 3 | 0 | 3 |
| HADH | SLC2A4 | 0.701016 | 4 | 0 | 4 |
| HADH | ARFGEF2 | 0.691014 | 3 | 0 | 3 |
| HADH | FRA10AC1 | 0.688856 | 3 | 0 | 3 |
| HADH | NMNAT1 | 0.680995 | 3 | 0 | 3 |
| HADH | NIPSNAP3A | 0.679321 | 4 | 0 | 4 |
| HADH | LRRC47 | 0.679301 | 4 | 0 | 4 |
| HADH | ZNF83 | 0.677729 | 3 | 0 | 3 |
| HADH | UQCR10 | 0.677336 | 4 | 0 | 4 |
| HADH | TMOD2 | 0.675326 | 4 | 0 | 4 |
| HADH | SRSF6 | 0.67512 | 4 | 0 | 3 |
| HADH | TADA2B | 0.67099 | 4 | 0 | 3 |
| HADH | BRWD1 | 0.669506 | 5 | 0 | 3 |
| HADH | TRA2B | 0.669038 | 4 | 0 | 3 |
For details and further investigation, click here