| Full name: histidine ammonia-lyase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q23.1 | ||
| Entrez ID: 3034 | HGNC ID: HGNC:4806 | Ensembl Gene: ENSG00000084110 | OMIM ID: 609457 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of HAL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HAL | 3034 | 217521_at | 0.1082 | 0.7179 | |
| GSE20347 | HAL | 3034 | 217521_at | -0.0823 | 0.4697 | |
| GSE23400 | HAL | 3034 | 217521_at | -0.1635 | 0.0001 | |
| GSE26886 | HAL | 3034 | 206643_at | -1.9871 | 0.0056 | |
| GSE29001 | HAL | 3034 | 217521_at | -0.1955 | 0.3125 | |
| GSE38129 | HAL | 3034 | 217521_at | -0.1866 | 0.0590 | |
| GSE45670 | HAL | 3034 | 206643_at | 0.5805 | 0.3111 | |
| GSE53622 | HAL | 3034 | 69226 | -0.7143 | 0.0014 | |
| GSE53624 | HAL | 3034 | 69226 | -1.0132 | 0.0000 | |
| GSE63941 | HAL | 3034 | 217521_at | 0.1923 | 0.2572 | |
| GSE77861 | HAL | 3034 | 217521_at | -0.0574 | 0.6147 | |
| SRP133303 | HAL | 3034 | RNAseq | -1.2391 | 0.0000 | |
| SRP193095 | HAL | 3034 | RNAseq | -0.4487 | 0.1128 | |
| TCGA | HAL | 3034 | RNAseq | 0.2425 | 0.6791 |
Upregulated datasets: 0; Downregulated datasets: 3.
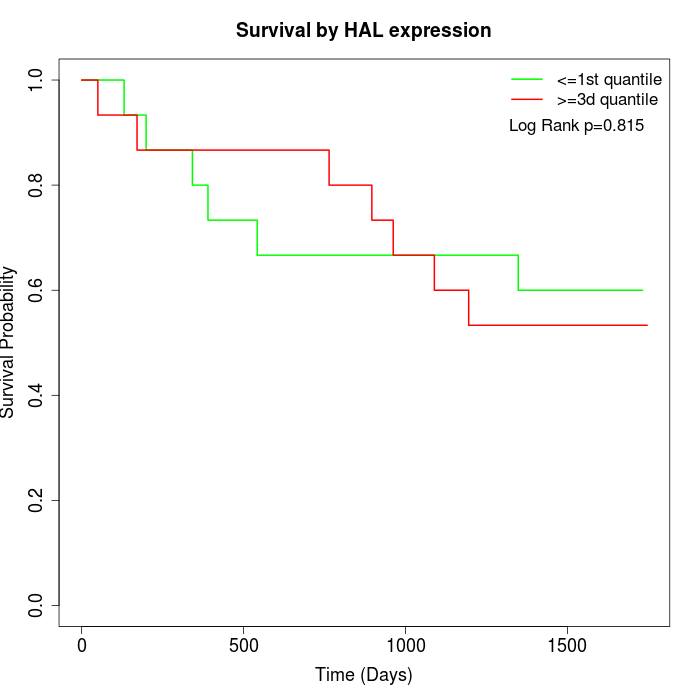
Survival by HAL expression:
Note: Click image to view full size file.
Copy number change of HAL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HAL | 3034 | 3 | 1 | 26 | |
| GSE20123 | HAL | 3034 | 3 | 1 | 26 | |
| GSE43470 | HAL | 3034 | 3 | 0 | 40 | |
| GSE46452 | HAL | 3034 | 9 | 1 | 49 | |
| GSE47630 | HAL | 3034 | 9 | 1 | 30 | |
| GSE54993 | HAL | 3034 | 0 | 5 | 65 | |
| GSE54994 | HAL | 3034 | 4 | 1 | 48 | |
| GSE60625 | HAL | 3034 | 0 | 0 | 11 | |
| GSE74703 | HAL | 3034 | 3 | 0 | 33 | |
| GSE74704 | HAL | 3034 | 1 | 1 | 18 | |
| TCGA | HAL | 3034 | 18 | 10 | 68 |
Total number of gains: 53; Total number of losses: 21; Total Number of normals: 414.
Somatic mutations of HAL:
Generating mutation plots.
Highly correlated genes for HAL:
Showing top 20/527 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HAL | CETN1 | 0.713822 | 5 | 0 | 4 |
| HAL | EDA2R | 0.70295 | 4 | 0 | 4 |
| HAL | GML | 0.700738 | 5 | 0 | 5 |
| HAL | CHST4 | 0.699128 | 4 | 0 | 4 |
| HAL | GABRB2 | 0.697871 | 6 | 0 | 5 |
| HAL | IFNA21 | 0.697605 | 4 | 0 | 4 |
| HAL | C3orf36 | 0.686073 | 4 | 0 | 4 |
| HAL | G6PC2 | 0.684983 | 4 | 0 | 4 |
| HAL | IL17A | 0.676984 | 4 | 0 | 4 |
| HAL | CER1 | 0.674962 | 5 | 0 | 5 |
| HAL | NCR2 | 0.674587 | 5 | 0 | 4 |
| HAL | CHAT | 0.667687 | 4 | 0 | 4 |
| HAL | P2RX6 | 0.666539 | 4 | 0 | 4 |
| HAL | SLC17A1 | 0.665974 | 5 | 0 | 4 |
| HAL | HHLA1 | 0.665835 | 4 | 0 | 4 |
| HAL | ITGB3 | 0.663347 | 3 | 0 | 3 |
| HAL | ATG10 | 0.662907 | 4 | 0 | 4 |
| HAL | PRL | 0.661016 | 4 | 0 | 4 |
| HAL | SLC22A13 | 0.660313 | 4 | 0 | 4 |
| HAL | BCAN | 0.659522 | 4 | 0 | 4 |
For details and further investigation, click here