| Full name: hes family bHLH transcription factor 7 | Alias Symbol: bHLHb37 | ||
| Type: protein-coding gene | Cytoband: 17p13.1 | ||
| Entrez ID: 84667 | HGNC ID: HGNC:15977 | Ensembl Gene: ENSG00000179111 | OMIM ID: 608059 |
| Drug and gene relationship at DGIdb | |||
Expression of HES7:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HES7 | 84667 | 224548_at | 0.1043 | 0.6008 | |
| GSE26886 | HES7 | 84667 | 224548_at | 0.3362 | 0.0085 | |
| GSE45670 | HES7 | 84667 | 224548_at | -0.1553 | 0.1284 | |
| GSE53622 | HES7 | 84667 | 87888 | 0.1050 | 0.5471 | |
| GSE53624 | HES7 | 84667 | 87888 | 1.0727 | 0.0000 | |
| GSE63941 | HES7 | 84667 | 224548_at | 0.3834 | 0.1900 | |
| GSE77861 | HES7 | 84667 | 224548_at | -0.1239 | 0.3042 | |
| SRP007169 | HES7 | 84667 | RNAseq | -2.8612 | 0.0008 | |
| SRP219564 | HES7 | 84667 | RNAseq | 1.9764 | 0.0004 | |
| TCGA | HES7 | 84667 | RNAseq | 2.7991 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 1.
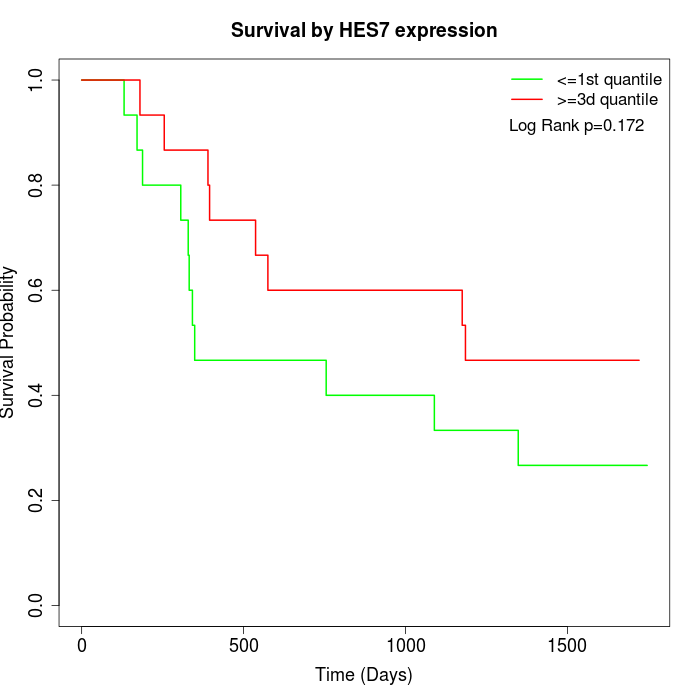
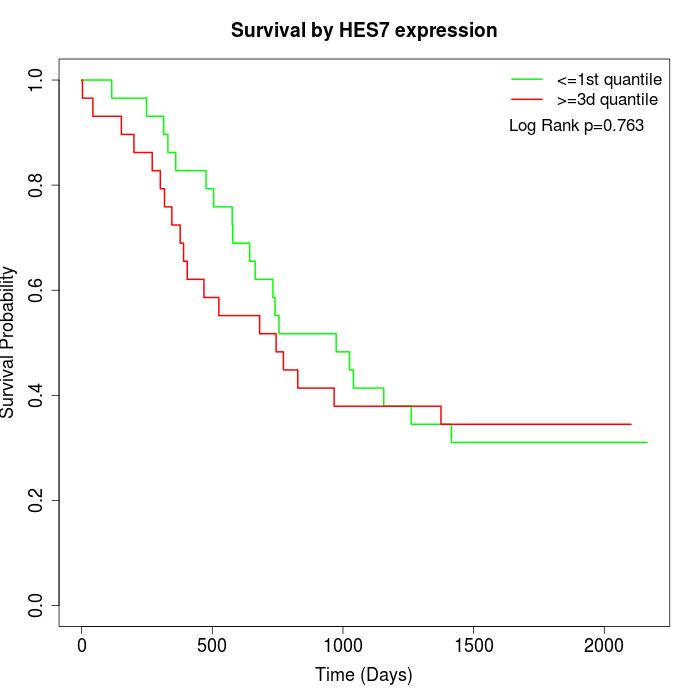
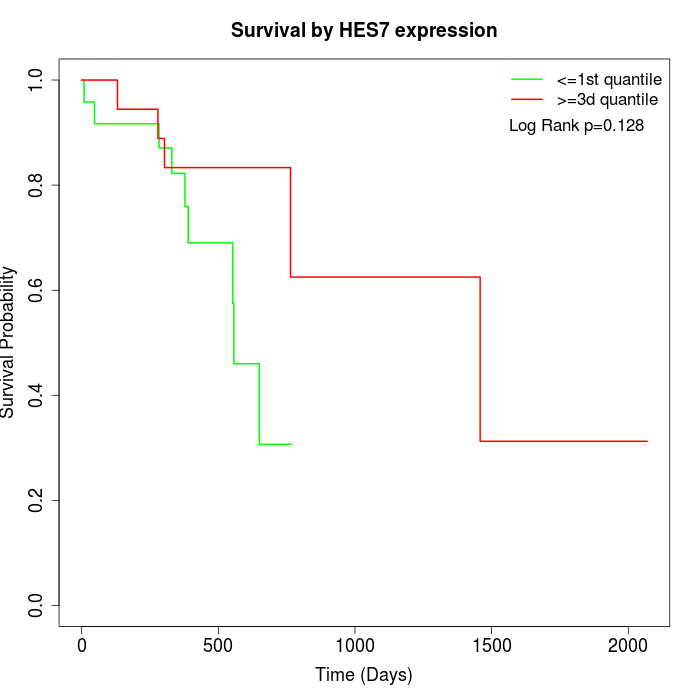
Survival by HES7 expression:
Note: Click image to view full size file.
Copy number change of HES7:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HES7 | 84667 | 4 | 2 | 24 | |
| GSE20123 | HES7 | 84667 | 4 | 3 | 23 | |
| GSE43470 | HES7 | 84667 | 1 | 6 | 36 | |
| GSE46452 | HES7 | 84667 | 34 | 1 | 24 | |
| GSE47630 | HES7 | 84667 | 7 | 1 | 32 | |
| GSE54993 | HES7 | 84667 | 4 | 3 | 63 | |
| GSE54994 | HES7 | 84667 | 5 | 8 | 40 | |
| GSE60625 | HES7 | 84667 | 4 | 0 | 7 | |
| GSE74703 | HES7 | 84667 | 1 | 3 | 32 | |
| GSE74704 | HES7 | 84667 | 2 | 1 | 17 | |
| TCGA | HES7 | 84667 | 17 | 21 | 58 |
Total number of gains: 83; Total number of losses: 49; Total Number of normals: 356.
Somatic mutations of HES7:
Generating mutation plots.
Highly correlated genes for HES7:
Showing top 20/36 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HES7 | BRF1 | 0.696833 | 3 | 0 | 3 |
| HES7 | KLRC3 | 0.693731 | 3 | 0 | 3 |
| HES7 | ARHGAP4 | 0.682756 | 3 | 0 | 3 |
| HES7 | HDAC8 | 0.672545 | 3 | 0 | 3 |
| HES7 | CD37 | 0.66228 | 3 | 0 | 3 |
| HES7 | TMEM51-AS1 | 0.656665 | 4 | 0 | 4 |
| HES7 | CPNE7 | 0.64813 | 4 | 0 | 3 |
| HES7 | PAX5 | 0.647939 | 3 | 0 | 3 |
| HES7 | AGAP2 | 0.629679 | 3 | 0 | 3 |
| HES7 | ARHGAP30 | 0.616713 | 3 | 0 | 3 |
| HES7 | WDR62 | 0.613815 | 3 | 0 | 3 |
| HES7 | IGLL1 | 0.604324 | 3 | 0 | 3 |
| HES7 | SLC39A5 | 0.59823 | 3 | 0 | 3 |
| HES7 | DLGAP2-AS1 | 0.592722 | 3 | 0 | 3 |
| HES7 | ZNF683 | 0.585613 | 4 | 0 | 3 |
| HES7 | LILRA1 | 0.584614 | 3 | 0 | 3 |
| HES7 | PRAM1 | 0.581196 | 4 | 0 | 3 |
| HES7 | LINC01010 | 0.579959 | 3 | 0 | 3 |
| HES7 | FMNL3 | 0.579015 | 4 | 0 | 3 |
| HES7 | PPP6R1 | 0.57256 | 3 | 0 | 3 |
For details and further investigation, click here