| Full name: Rho GTPase activating protein 4 | Alias Symbol: KIAA0131|C1|p115|RhoGAP4|SrGAP4 | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 393 | HGNC ID: HGNC:674 | Ensembl Gene: ENSG00000089820 | OMIM ID: 300023 |
| Drug and gene relationship at DGIdb | |||
Expression of ARHGAP4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGAP4 | 393 | 204425_at | 0.3775 | 0.5067 | |
| GSE20347 | ARHGAP4 | 393 | 204425_at | 0.3013 | 0.0329 | |
| GSE23400 | ARHGAP4 | 393 | 204425_at | 0.1058 | 0.0668 | |
| GSE26886 | ARHGAP4 | 393 | 204425_at | 0.4114 | 0.0168 | |
| GSE29001 | ARHGAP4 | 393 | 204425_at | 0.4640 | 0.0438 | |
| GSE38129 | ARHGAP4 | 393 | 204425_at | 0.1249 | 0.4337 | |
| GSE45670 | ARHGAP4 | 393 | 204425_at | -0.0304 | 0.7953 | |
| GSE53622 | ARHGAP4 | 393 | 96941 | -0.1974 | 0.0008 | |
| GSE53624 | ARHGAP4 | 393 | 96941 | -0.2545 | 0.0000 | |
| GSE63941 | ARHGAP4 | 393 | 204425_at | 0.2313 | 0.4843 | |
| GSE77861 | ARHGAP4 | 393 | 204425_at | 0.0040 | 0.9842 | |
| GSE97050 | ARHGAP4 | 393 | A_33_P3282181 | -0.2348 | 0.3972 | |
| SRP007169 | ARHGAP4 | 393 | RNAseq | 0.8240 | 0.1891 | |
| SRP008496 | ARHGAP4 | 393 | RNAseq | 0.1700 | 0.6510 | |
| SRP064894 | ARHGAP4 | 393 | RNAseq | 0.8906 | 0.0078 | |
| SRP133303 | ARHGAP4 | 393 | RNAseq | -0.1248 | 0.5761 | |
| SRP159526 | ARHGAP4 | 393 | RNAseq | -0.7439 | 0.1099 | |
| SRP193095 | ARHGAP4 | 393 | RNAseq | 0.5329 | 0.0706 | |
| SRP219564 | ARHGAP4 | 393 | RNAseq | 0.6124 | 0.0839 | |
| TCGA | ARHGAP4 | 393 | RNAseq | 0.0055 | 0.9650 |
Upregulated datasets: 0; Downregulated datasets: 0.
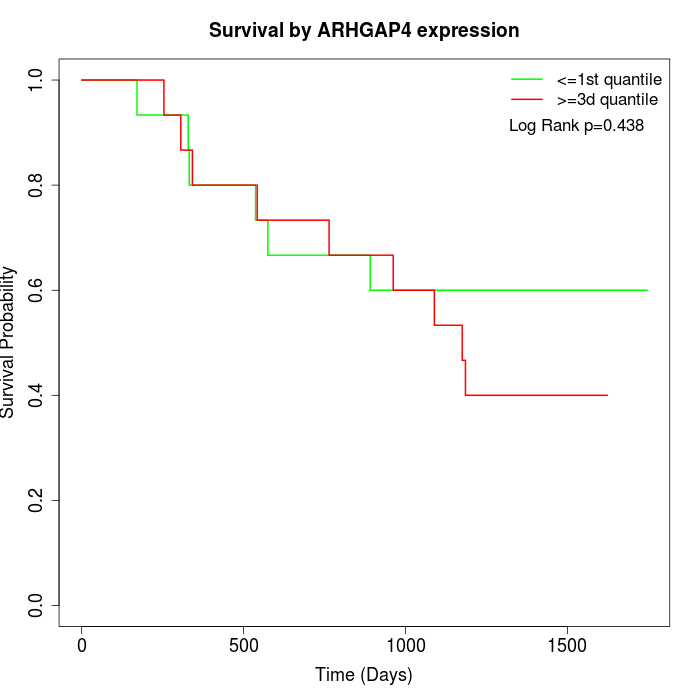
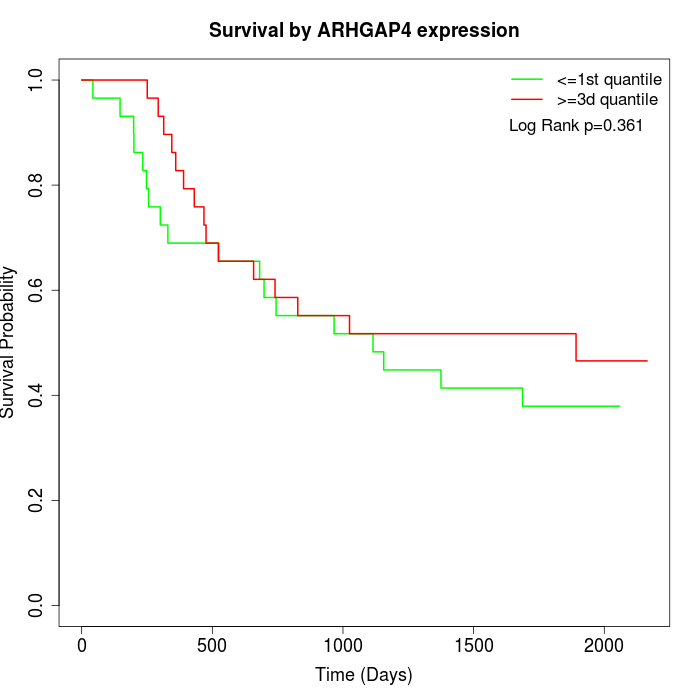
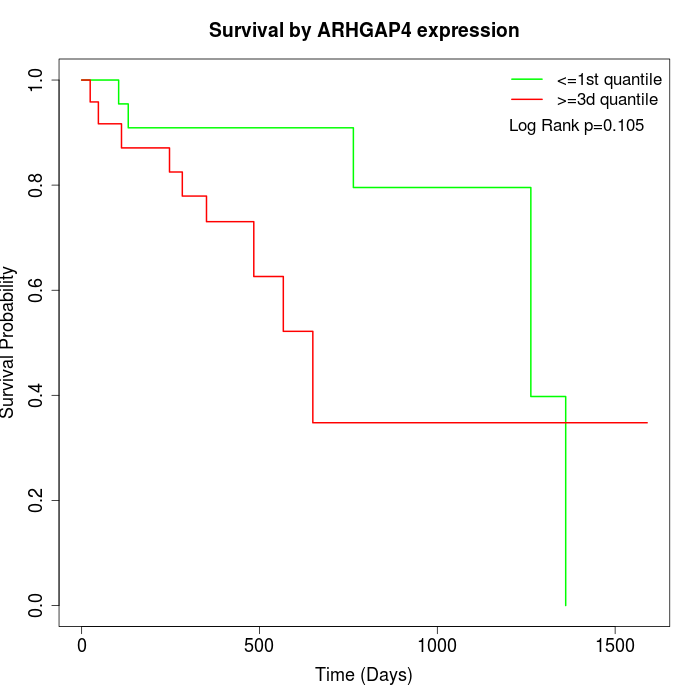
Survival by ARHGAP4 expression:
Note: Click image to view full size file.
Copy number change of ARHGAP4:
No record found for this gene.
Somatic mutations of ARHGAP4:
Generating mutation plots.
Highly correlated genes for ARHGAP4:
Showing top 20/376 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGAP4 | ZNF683 | 0.762942 | 4 | 0 | 4 |
| ARHGAP4 | CTXN1 | 0.750545 | 3 | 0 | 3 |
| ARHGAP4 | TBC1D25 | 0.743151 | 3 | 0 | 3 |
| ARHGAP4 | FILIP1 | 0.726827 | 4 | 0 | 4 |
| ARHGAP4 | DIRAS1 | 0.72374 | 4 | 0 | 4 |
| ARHGAP4 | MTMR11 | 0.719056 | 3 | 0 | 3 |
| ARHGAP4 | MAPRE3 | 0.71358 | 4 | 0 | 3 |
| ARHGAP4 | TEAD2 | 0.712862 | 4 | 0 | 3 |
| ARHGAP4 | ELFN1 | 0.699933 | 5 | 0 | 5 |
| ARHGAP4 | CELA3A | 0.698458 | 3 | 0 | 3 |
| ARHGAP4 | RAB39B | 0.698029 | 3 | 0 | 3 |
| ARHGAP4 | KIAA0513 | 0.696331 | 3 | 0 | 3 |
| ARHGAP4 | SMYD1 | 0.692834 | 3 | 0 | 3 |
| ARHGAP4 | IL18BP | 0.690912 | 3 | 0 | 3 |
| ARHGAP4 | GPR3 | 0.690286 | 3 | 0 | 3 |
| ARHGAP4 | SLC39A3 | 0.6873 | 4 | 0 | 4 |
| ARHGAP4 | NME4 | 0.686143 | 3 | 0 | 3 |
| ARHGAP4 | C2CD4B | 0.685009 | 3 | 0 | 3 |
| ARHGAP4 | HES7 | 0.682756 | 3 | 0 | 3 |
| ARHGAP4 | SSBP4 | 0.682733 | 3 | 0 | 3 |
For details and further investigation, click here