| Full name: long intergenic non-protein coding RNA 1010 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 6q23.2 | ||
| Entrez ID: 154092 | HGNC ID: HGNC:48978 | Ensembl Gene: ENSG00000236700 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC01010:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC01010 | 154092 | 237731_at | 0.2669 | 0.5144 | |
| GSE26886 | LINC01010 | 154092 | 237731_at | 0.5439 | 0.0031 | |
| GSE45670 | LINC01010 | 154092 | 237731_at | 0.4364 | 0.0004 | |
| GSE53622 | LINC01010 | 154092 | 44229 | 1.2557 | 0.0000 | |
| GSE53624 | LINC01010 | 154092 | 44229 | 1.7416 | 0.0000 | |
| GSE63941 | LINC01010 | 154092 | 237731_at | 0.2071 | 0.3057 | |
| GSE77861 | LINC01010 | 154092 | 237731_at | 0.0287 | 0.8721 | |
| SRP133303 | LINC01010 | 154092 | RNAseq | 1.3692 | 0.0054 | |
| SRP159526 | LINC01010 | 154092 | RNAseq | 2.9682 | 0.0003 |
Upregulated datasets: 4; Downregulated datasets: 0.
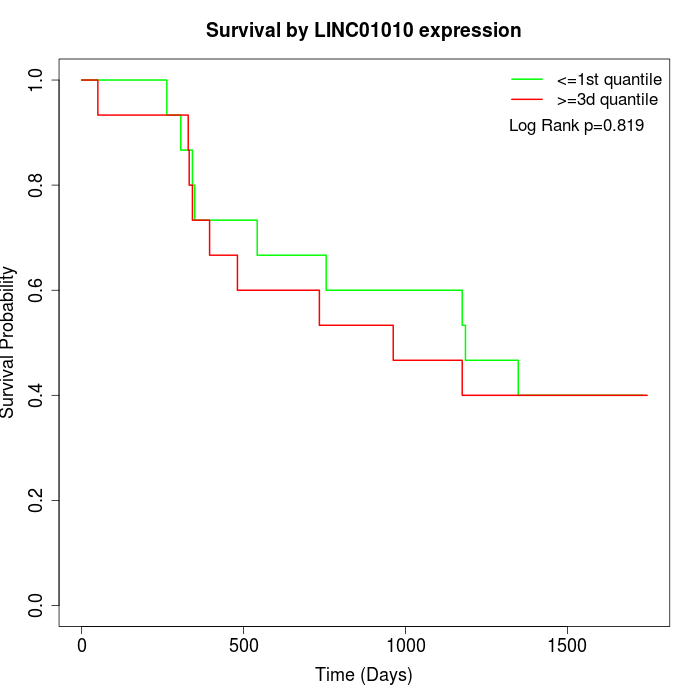
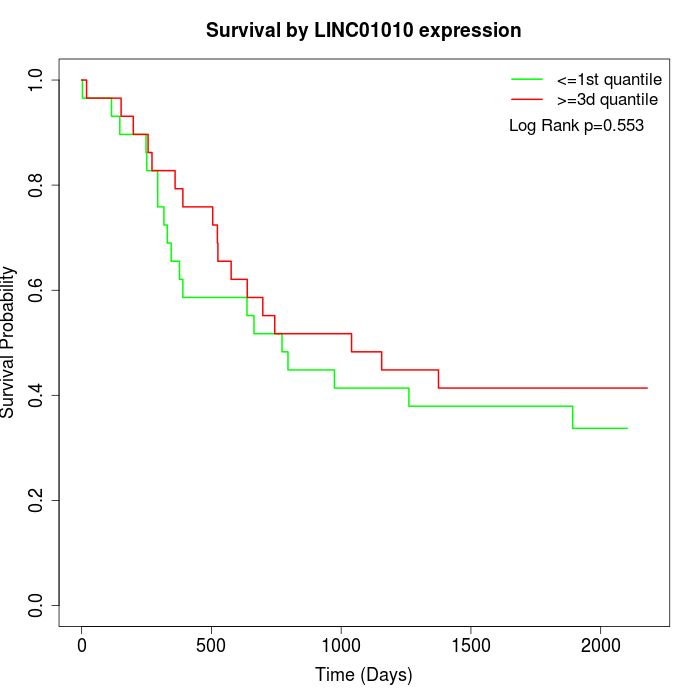
Survival by LINC01010 expression:
Note: Click image to view full size file.
Copy number change of LINC01010:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC01010 | 154092 | 5 | 4 | 21 | |
| GSE20123 | LINC01010 | 154092 | 5 | 3 | 22 | |
| GSE43470 | LINC01010 | 154092 | 5 | 0 | 38 | |
| GSE46452 | LINC01010 | 154092 | 3 | 10 | 46 | |
| GSE47630 | LINC01010 | 154092 | 11 | 4 | 25 | |
| GSE54993 | LINC01010 | 154092 | 3 | 2 | 65 | |
| GSE54994 | LINC01010 | 154092 | 10 | 7 | 36 | |
| GSE60625 | LINC01010 | 154092 | 0 | 1 | 10 | |
| GSE74703 | LINC01010 | 154092 | 4 | 0 | 32 | |
| GSE74704 | LINC01010 | 154092 | 2 | 1 | 17 |
Total number of gains: 48; Total number of losses: 32; Total Number of normals: 312.
Somatic mutations of LINC01010:
Generating mutation plots.
Highly correlated genes for LINC01010:
Showing top 20/81 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC01010 | DNLZ | 0.697711 | 3 | 0 | 3 |
| LINC01010 | TP53I11 | 0.697076 | 3 | 0 | 3 |
| LINC01010 | CDH4 | 0.686461 | 3 | 0 | 3 |
| LINC01010 | FBN3 | 0.685597 | 3 | 0 | 3 |
| LINC01010 | TMEM132A | 0.682714 | 3 | 0 | 3 |
| LINC01010 | NTSR2 | 0.680096 | 3 | 0 | 3 |
| LINC01010 | LIN37 | 0.673227 | 3 | 0 | 3 |
| LINC01010 | ATP2B2 | 0.669005 | 4 | 0 | 3 |
| LINC01010 | RHPN1 | 0.668866 | 3 | 0 | 3 |
| LINC01010 | PLEKHA4 | 0.657075 | 3 | 0 | 3 |
| LINC01010 | HGD | 0.653499 | 3 | 0 | 3 |
| LINC01010 | SLC29A4 | 0.652107 | 3 | 0 | 3 |
| LINC01010 | FRS3 | 0.644532 | 3 | 0 | 3 |
| LINC01010 | CEMP1 | 0.64123 | 3 | 0 | 3 |
| LINC01010 | AGAP2 | 0.639709 | 4 | 0 | 4 |
| LINC01010 | QSOX2 | 0.631193 | 3 | 0 | 3 |
| LINC01010 | ZNF787 | 0.630324 | 3 | 0 | 3 |
| LINC01010 | MAP2K7 | 0.62941 | 3 | 0 | 3 |
| LINC01010 | PHOX2A | 0.621176 | 3 | 0 | 3 |
| LINC01010 | IL18BP | 0.620336 | 3 | 0 | 3 |
For details and further investigation, click here