| Full name: hexokinase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q22.1 | ||
| Entrez ID: 3098 | HGNC ID: HGNC:4922 | Ensembl Gene: ENSG00000156515 | OMIM ID: 142600 |
| Drug and gene relationship at DGIdb | |||
HK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa04930 | Type II diabetes mellitus |
Expression of HK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HK1 | 3098 | 200697_at | -0.3489 | 0.3012 | |
| GSE20347 | HK1 | 3098 | 200697_at | -0.5003 | 0.0015 | |
| GSE23400 | HK1 | 3098 | 200697_at | -0.3886 | 0.0000 | |
| GSE26886 | HK1 | 3098 | 200697_at | -0.4987 | 0.0075 | |
| GSE29001 | HK1 | 3098 | 200697_at | -0.6131 | 0.0011 | |
| GSE38129 | HK1 | 3098 | 200697_at | -0.2988 | 0.0605 | |
| GSE45670 | HK1 | 3098 | 200697_at | -0.2498 | 0.0883 | |
| GSE53622 | HK1 | 3098 | 47701 | -0.7662 | 0.0000 | |
| GSE53624 | HK1 | 3098 | 47701 | -0.5531 | 0.0000 | |
| GSE63941 | HK1 | 3098 | 200697_at | 0.1514 | 0.7802 | |
| GSE77861 | HK1 | 3098 | 200697_at | -0.1615 | 0.5724 | |
| GSE97050 | HK1 | 3098 | A_23_P217958 | -0.4800 | 0.1476 | |
| SRP007169 | HK1 | 3098 | RNAseq | 0.2487 | 0.5692 | |
| SRP008496 | HK1 | 3098 | RNAseq | 0.5246 | 0.0908 | |
| SRP064894 | HK1 | 3098 | RNAseq | -0.9640 | 0.0000 | |
| SRP133303 | HK1 | 3098 | RNAseq | -0.5946 | 0.0003 | |
| SRP159526 | HK1 | 3098 | RNAseq | -0.4612 | 0.3800 | |
| SRP193095 | HK1 | 3098 | RNAseq | -0.4998 | 0.0080 | |
| SRP219564 | HK1 | 3098 | RNAseq | -0.7216 | 0.0017 | |
| TCGA | HK1 | 3098 | RNAseq | -0.0470 | 0.4246 |
Upregulated datasets: 0; Downregulated datasets: 0.
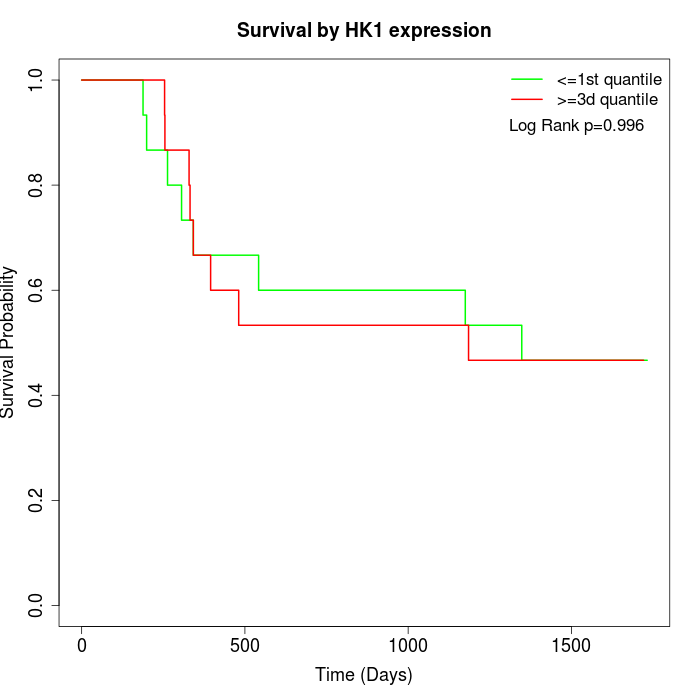
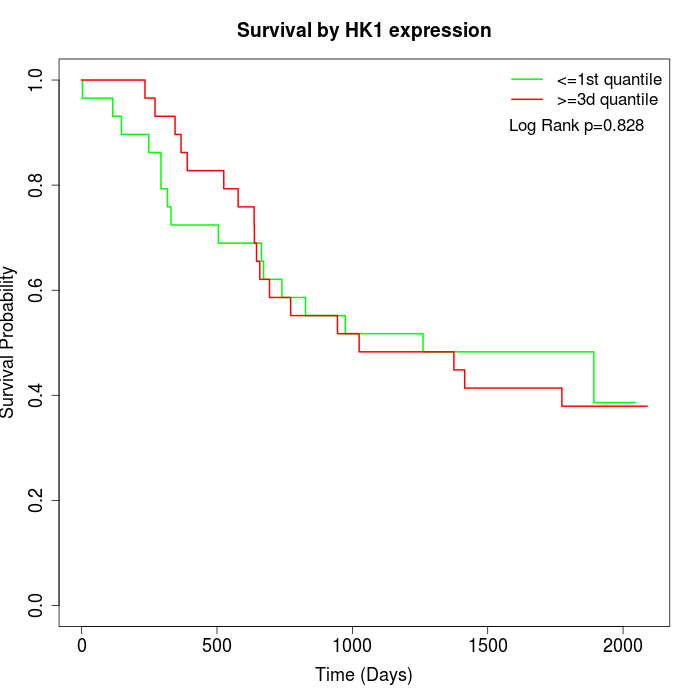
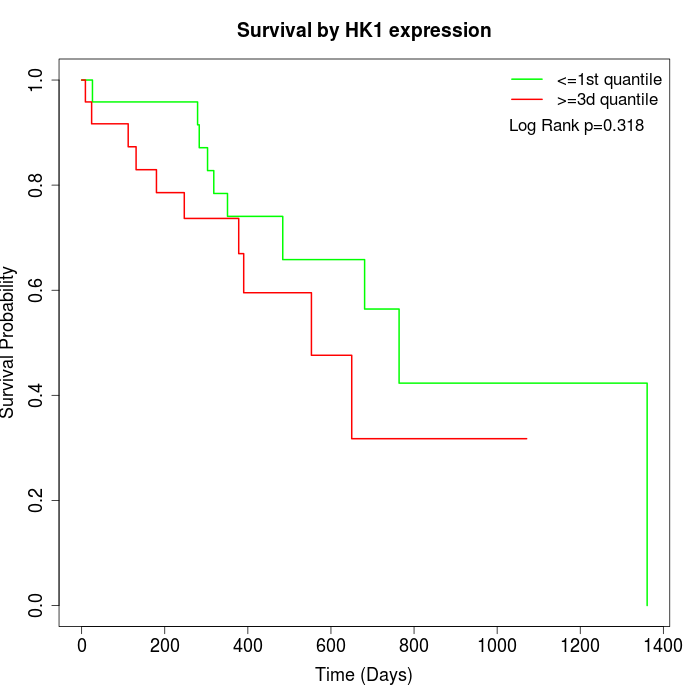
Survival by HK1 expression:
Note: Click image to view full size file.
Copy number change of HK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HK1 | 3098 | 3 | 4 | 23 | |
| GSE20123 | HK1 | 3098 | 3 | 3 | 24 | |
| GSE43470 | HK1 | 3098 | 1 | 7 | 35 | |
| GSE46452 | HK1 | 3098 | 0 | 11 | 48 | |
| GSE47630 | HK1 | 3098 | 2 | 14 | 24 | |
| GSE54993 | HK1 | 3098 | 7 | 0 | 63 | |
| GSE54994 | HK1 | 3098 | 2 | 12 | 39 | |
| GSE60625 | HK1 | 3098 | 0 | 0 | 11 | |
| GSE74703 | HK1 | 3098 | 1 | 4 | 31 | |
| GSE74704 | HK1 | 3098 | 1 | 1 | 18 | |
| TCGA | HK1 | 3098 | 10 | 23 | 63 |
Total number of gains: 30; Total number of losses: 79; Total Number of normals: 379.
Somatic mutations of HK1:
Generating mutation plots.
Highly correlated genes for HK1:
Showing top 20/1091 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HK1 | FBXW5 | 0.813729 | 3 | 0 | 3 |
| HK1 | IDH3G | 0.806946 | 3 | 0 | 3 |
| HK1 | BAGE | 0.769285 | 3 | 0 | 3 |
| HK1 | HKDC1 | 0.761982 | 4 | 0 | 4 |
| HK1 | RAB4B | 0.7602 | 3 | 0 | 3 |
| HK1 | TOM1L1 | 0.752875 | 3 | 0 | 3 |
| HK1 | RNF207 | 0.749311 | 3 | 0 | 3 |
| HK1 | TCEANC2 | 0.736878 | 3 | 0 | 3 |
| HK1 | APBA1 | 0.731702 | 3 | 0 | 3 |
| HK1 | CD99 | 0.726211 | 3 | 0 | 3 |
| HK1 | LIPF | 0.723099 | 3 | 0 | 3 |
| HK1 | CPEB4 | 0.720511 | 4 | 0 | 3 |
| HK1 | SH2B1 | 0.716151 | 3 | 0 | 3 |
| HK1 | PTGR1 | 0.714409 | 8 | 0 | 7 |
| HK1 | CNST | 0.714278 | 4 | 0 | 4 |
| HK1 | ZNF528 | 0.703548 | 3 | 0 | 3 |
| HK1 | TWF2 | 0.701764 | 4 | 0 | 4 |
| HK1 | ALDH3A1 | 0.700251 | 11 | 0 | 11 |
| HK1 | CYP4F3 | 0.697167 | 11 | 0 | 10 |
| HK1 | NLRX1 | 0.692134 | 10 | 0 | 10 |
For details and further investigation, click here