| Full name: NLR family member X1 | Alias Symbol: NOD9|CLR11.3 | ||
| Type: protein-coding gene | Cytoband: 11q23.3 | ||
| Entrez ID: 79671 | HGNC ID: HGNC:29890 | Ensembl Gene: ENSG00000160703 | OMIM ID: 611947 |
| Drug and gene relationship at DGIdb | |||
NLRX1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04622 | RIG-I-like receptor signaling pathway |
Expression of NLRX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NLRX1 | 79671 | 219680_at | -1.3127 | 0.0567 | |
| GSE20347 | NLRX1 | 79671 | 219680_at | -1.1028 | 0.0000 | |
| GSE23400 | NLRX1 | 79671 | 219680_at | -0.8645 | 0.0000 | |
| GSE26886 | NLRX1 | 79671 | 219680_at | -1.4339 | 0.0000 | |
| GSE29001 | NLRX1 | 79671 | 219680_at | -1.2464 | 0.0000 | |
| GSE38129 | NLRX1 | 79671 | 219680_at | -0.9615 | 0.0000 | |
| GSE45670 | NLRX1 | 79671 | 219680_at | -0.2555 | 0.3133 | |
| GSE53622 | NLRX1 | 79671 | 121679 | -1.5602 | 0.0000 | |
| GSE53624 | NLRX1 | 79671 | 121679 | -1.6938 | 0.0000 | |
| GSE63941 | NLRX1 | 79671 | 219680_at | -0.3540 | 0.3183 | |
| GSE77861 | NLRX1 | 79671 | 219680_at | -0.5040 | 0.1287 | |
| GSE97050 | NLRX1 | 79671 | A_24_P244356 | -0.6316 | 0.1195 | |
| SRP007169 | NLRX1 | 79671 | RNAseq | -2.9901 | 0.0000 | |
| SRP008496 | NLRX1 | 79671 | RNAseq | -3.0945 | 0.0000 | |
| SRP064894 | NLRX1 | 79671 | RNAseq | -1.4453 | 0.0000 | |
| SRP133303 | NLRX1 | 79671 | RNAseq | -1.6163 | 0.0000 | |
| SRP159526 | NLRX1 | 79671 | RNAseq | -2.1148 | 0.0000 | |
| SRP193095 | NLRX1 | 79671 | RNAseq | -1.9295 | 0.0000 | |
| SRP219564 | NLRX1 | 79671 | RNAseq | -2.1874 | 0.0000 | |
| TCGA | NLRX1 | 79671 | RNAseq | -0.0429 | 0.6465 |
Upregulated datasets: 0; Downregulated datasets: 12.
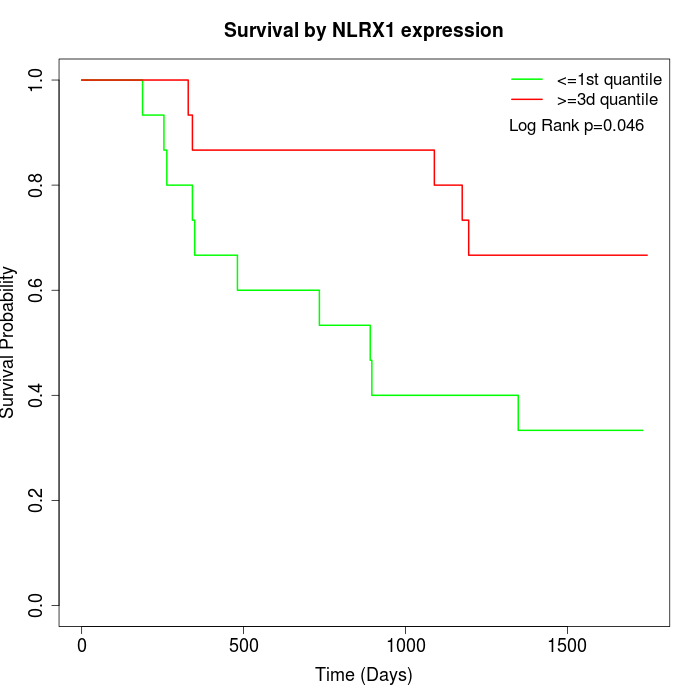
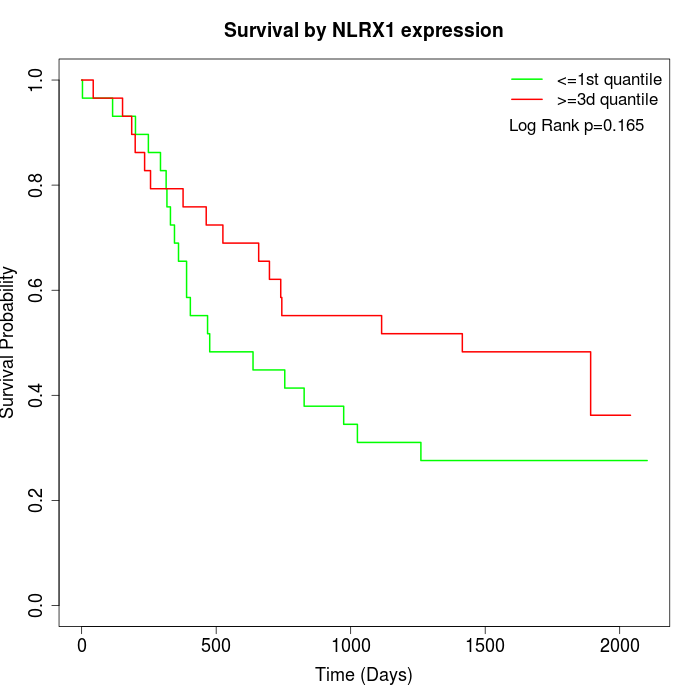
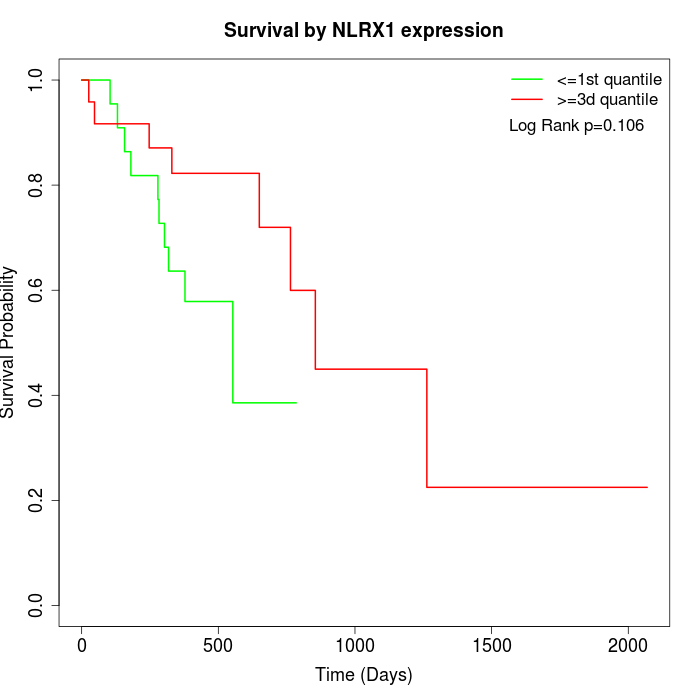
Survival by NLRX1 expression:
Note: Click image to view full size file.
Copy number change of NLRX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NLRX1 | 79671 | 1 | 11 | 18 | |
| GSE20123 | NLRX1 | 79671 | 1 | 11 | 18 | |
| GSE43470 | NLRX1 | 79671 | 2 | 8 | 33 | |
| GSE46452 | NLRX1 | 79671 | 3 | 26 | 30 | |
| GSE47630 | NLRX1 | 79671 | 2 | 19 | 19 | |
| GSE54993 | NLRX1 | 79671 | 10 | 0 | 60 | |
| GSE54994 | NLRX1 | 79671 | 5 | 19 | 29 | |
| GSE60625 | NLRX1 | 79671 | 0 | 3 | 8 | |
| GSE74703 | NLRX1 | 79671 | 1 | 5 | 30 | |
| GSE74704 | NLRX1 | 79671 | 0 | 7 | 13 | |
| TCGA | NLRX1 | 79671 | 4 | 51 | 41 |
Total number of gains: 29; Total number of losses: 160; Total Number of normals: 299.
Somatic mutations of NLRX1:
Generating mutation plots.
Highly correlated genes for NLRX1:
Showing top 20/1720 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NLRX1 | LRP10 | 0.825506 | 10 | 0 | 10 |
| NLRX1 | TOM1 | 0.815357 | 11 | 0 | 10 |
| NLRX1 | PIM1 | 0.813456 | 10 | 0 | 10 |
| NLRX1 | CRCT1 | 0.809271 | 10 | 0 | 10 |
| NLRX1 | EVPL | 0.808609 | 11 | 0 | 11 |
| NLRX1 | SCNN1B | 0.806777 | 10 | 0 | 10 |
| NLRX1 | ULK3 | 0.806574 | 7 | 0 | 7 |
| NLRX1 | PLEKHM1 | 0.799143 | 12 | 0 | 11 |
| NLRX1 | SLC16A6 | 0.7982 | 11 | 0 | 11 |
| NLRX1 | MXD1 | 0.793477 | 10 | 0 | 9 |
| NLRX1 | CLTB | 0.793315 | 10 | 0 | 10 |
| NLRX1 | ANKRD37 | 0.792963 | 4 | 0 | 4 |
| NLRX1 | CHP1 | 0.792723 | 10 | 0 | 10 |
| NLRX1 | ENDOU | 0.792064 | 11 | 0 | 10 |
| NLRX1 | PPL | 0.791085 | 11 | 0 | 10 |
| NLRX1 | LDHD | 0.791058 | 6 | 0 | 6 |
| NLRX1 | KLK13 | 0.789217 | 11 | 0 | 10 |
| NLRX1 | ANXA9 | 0.784733 | 11 | 0 | 11 |
| NLRX1 | MGLL | 0.784437 | 11 | 0 | 11 |
| NLRX1 | RMND5B | 0.784421 | 11 | 0 | 11 |
For details and further investigation, click here