| Full name: 3-hydroxy-3-methylglutaryl-CoA reductase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5q13.3 | ||
| Entrez ID: 3156 | HGNC ID: HGNC:5006 | Ensembl Gene: ENSG00000113161 | OMIM ID: 142910 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
HMGCR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04152 | AMPK signaling pathway |
Expression of HMGCR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HMGCR | 3156 | 202540_s_at | -0.2581 | 0.8107 | |
| GSE20347 | HMGCR | 3156 | 202540_s_at | -0.9999 | 0.0000 | |
| GSE23400 | HMGCR | 3156 | 202540_s_at | -0.5481 | 0.0000 | |
| GSE26886 | HMGCR | 3156 | 202540_s_at | -1.3124 | 0.0000 | |
| GSE29001 | HMGCR | 3156 | 202540_s_at | -0.8508 | 0.0014 | |
| GSE38129 | HMGCR | 3156 | 202540_s_at | -0.3950 | 0.1285 | |
| GSE45670 | HMGCR | 3156 | 202540_s_at | 0.0110 | 0.9609 | |
| GSE53622 | HMGCR | 3156 | 6790 | -0.6813 | 0.0000 | |
| GSE53624 | HMGCR | 3156 | 6790 | -0.8898 | 0.0000 | |
| GSE63941 | HMGCR | 3156 | 202539_s_at | 0.0827 | 0.8932 | |
| GSE77861 | HMGCR | 3156 | 202540_s_at | -0.6121 | 0.0359 | |
| GSE97050 | HMGCR | 3156 | A_23_P30495 | 0.0625 | 0.8138 | |
| SRP007169 | HMGCR | 3156 | RNAseq | -0.8906 | 0.0035 | |
| SRP008496 | HMGCR | 3156 | RNAseq | -1.1279 | 0.0000 | |
| SRP064894 | HMGCR | 3156 | RNAseq | -1.5941 | 0.0000 | |
| SRP133303 | HMGCR | 3156 | RNAseq | -0.8655 | 0.0009 | |
| SRP159526 | HMGCR | 3156 | RNAseq | -0.7034 | 0.0741 | |
| SRP193095 | HMGCR | 3156 | RNAseq | -1.2944 | 0.0000 | |
| SRP219564 | HMGCR | 3156 | RNAseq | -1.4845 | 0.0149 | |
| TCGA | HMGCR | 3156 | RNAseq | -0.0020 | 0.9786 |
Upregulated datasets: 0; Downregulated datasets: 5.
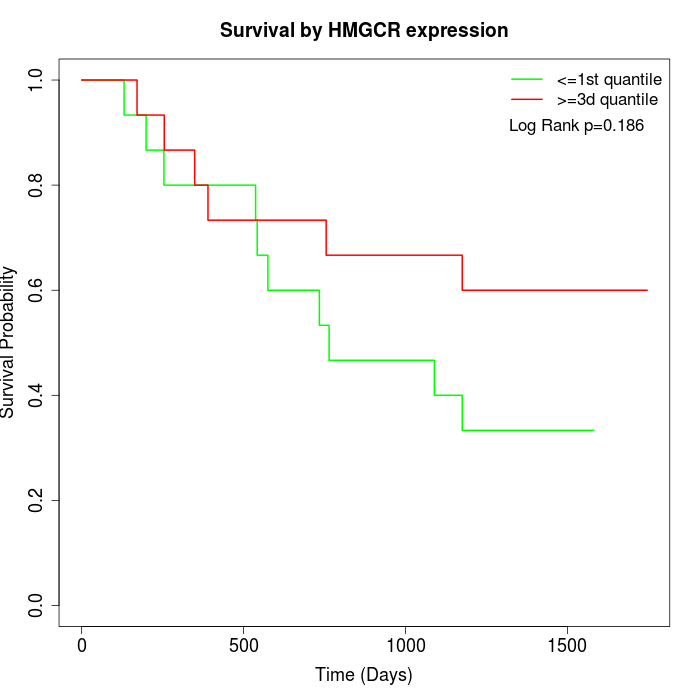
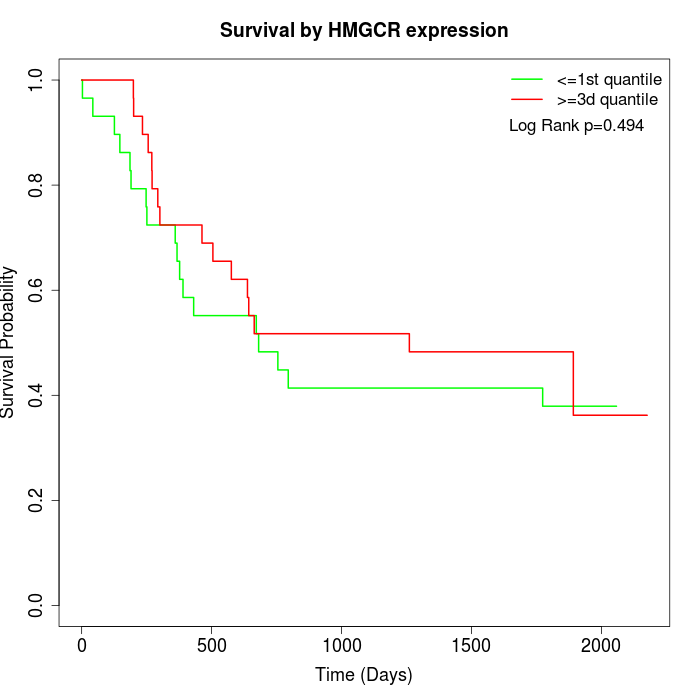
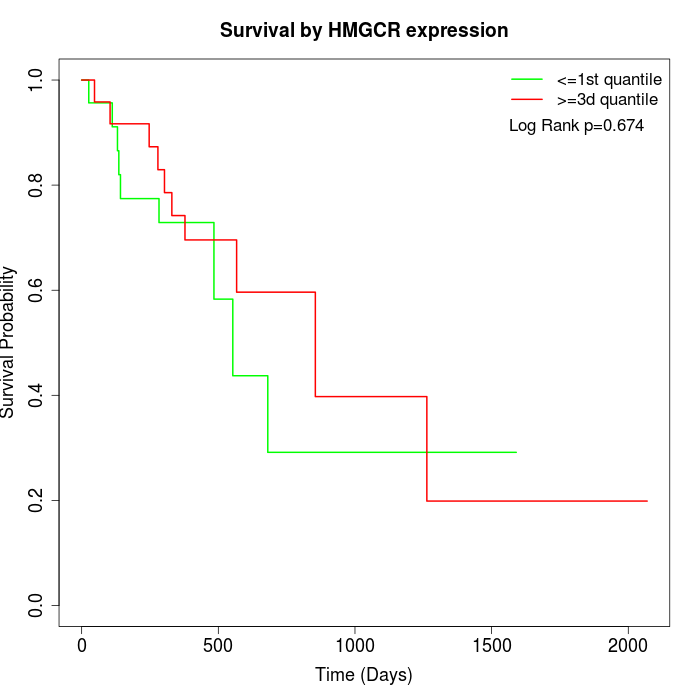
Survival by HMGCR expression:
Note: Click image to view full size file.
Copy number change of HMGCR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HMGCR | 3156 | 1 | 13 | 16 | |
| GSE20123 | HMGCR | 3156 | 1 | 13 | 16 | |
| GSE43470 | HMGCR | 3156 | 1 | 13 | 29 | |
| GSE46452 | HMGCR | 3156 | 0 | 28 | 31 | |
| GSE47630 | HMGCR | 3156 | 1 | 19 | 20 | |
| GSE54993 | HMGCR | 3156 | 8 | 1 | 61 | |
| GSE54994 | HMGCR | 3156 | 1 | 19 | 33 | |
| GSE60625 | HMGCR | 3156 | 0 | 0 | 11 | |
| GSE74703 | HMGCR | 3156 | 1 | 9 | 26 | |
| GSE74704 | HMGCR | 3156 | 1 | 6 | 13 | |
| TCGA | HMGCR | 3156 | 4 | 50 | 42 |
Total number of gains: 19; Total number of losses: 171; Total Number of normals: 298.
Somatic mutations of HMGCR:
Generating mutation plots.
Highly correlated genes for HMGCR:
Showing top 20/1276 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HMGCR | MSMO1 | 0.763222 | 12 | 0 | 12 |
| HMGCR | LYSMD3 | 0.747316 | 4 | 0 | 4 |
| HMGCR | CCNYL1 | 0.73544 | 4 | 0 | 4 |
| HMGCR | CWH43 | 0.724269 | 9 | 0 | 9 |
| HMGCR | TMEM79 | 0.722964 | 7 | 0 | 7 |
| HMGCR | SH3PXD2A-AS1 | 0.722961 | 3 | 0 | 3 |
| HMGCR | C15orf48 | 0.722709 | 6 | 0 | 6 |
| HMGCR | CAPN14 | 0.72238 | 5 | 0 | 5 |
| HMGCR | MREG | 0.718694 | 11 | 0 | 11 |
| HMGCR | ZNF185 | 0.709732 | 11 | 0 | 10 |
| HMGCR | CDS1 | 0.708608 | 12 | 0 | 10 |
| HMGCR | FRMD4B | 0.70723 | 10 | 0 | 8 |
| HMGCR | VPS4B | 0.707205 | 10 | 0 | 9 |
| HMGCR | DMKN | 0.706427 | 6 | 0 | 5 |
| HMGCR | TMEM40 | 0.70335 | 11 | 0 | 10 |
| HMGCR | C1orf210 | 0.699254 | 7 | 0 | 7 |
| HMGCR | RDH12 | 0.69901 | 7 | 0 | 7 |
| HMGCR | IPMK | 0.697245 | 4 | 0 | 3 |
| HMGCR | ACPP | 0.697015 | 11 | 0 | 10 |
| HMGCR | A2ML1 | 0.694253 | 6 | 0 | 6 |
For details and further investigation, click here