| Full name: heat shock protein family B (small) member 1 | Alias Symbol: HSP27|HSP28|Hs.76067|Hsp25|CMT2F | ||
| Type: protein-coding gene | Cytoband: 7q11.23 | ||
| Entrez ID: 3315 | HGNC ID: HGNC:5246 | Ensembl Gene: ENSG00000106211 | OMIM ID: 602195 |
| Drug and gene relationship at DGIdb | |||
HSPB1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04370 | VEGF signaling pathway |
Expression of HSPB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HSPB1 | 3315 | 201841_s_at | -0.0212 | 0.9538 | |
| GSE20347 | HSPB1 | 3315 | 201841_s_at | -0.4107 | 0.0060 | |
| GSE23400 | HSPB1 | 3315 | 201841_s_at | -0.6259 | 0.0000 | |
| GSE26886 | HSPB1 | 3315 | 201841_s_at | -1.1844 | 0.0234 | |
| GSE29001 | HSPB1 | 3315 | 201841_s_at | -0.8169 | 0.0397 | |
| GSE38129 | HSPB1 | 3315 | 201841_s_at | -0.2015 | 0.3507 | |
| GSE45670 | HSPB1 | 3315 | 201841_s_at | 0.0666 | 0.7348 | |
| GSE53622 | HSPB1 | 3315 | 59105 | -1.4180 | 0.0000 | |
| GSE53624 | HSPB1 | 3315 | 59105 | -0.7522 | 0.0000 | |
| GSE63941 | HSPB1 | 3315 | 201841_s_at | 1.8900 | 0.3030 | |
| GSE77861 | HSPB1 | 3315 | 201841_s_at | -0.2282 | 0.4471 | |
| GSE97050 | HSPB1 | A_21_P0013662 | -0.2060 | 0.6783 | ||
| SRP007169 | HSPB1 | 3315 | RNAseq | -1.7862 | 0.0000 | |
| SRP008496 | HSPB1 | 3315 | RNAseq | -1.5165 | 0.0000 | |
| SRP064894 | HSPB1 | 3315 | RNAseq | -0.1880 | 0.6284 | |
| SRP133303 | HSPB1 | 3315 | RNAseq | -0.5467 | 0.0788 | |
| SRP159526 | HSPB1 | 3315 | RNAseq | -1.0469 | 0.0269 | |
| SRP193095 | HSPB1 | 3315 | RNAseq | -0.8992 | 0.0083 | |
| SRP219564 | HSPB1 | 3315 | RNAseq | -0.7500 | 0.0922 | |
| TCGA | HSPB1 | 3315 | RNAseq | 0.3770 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 5.
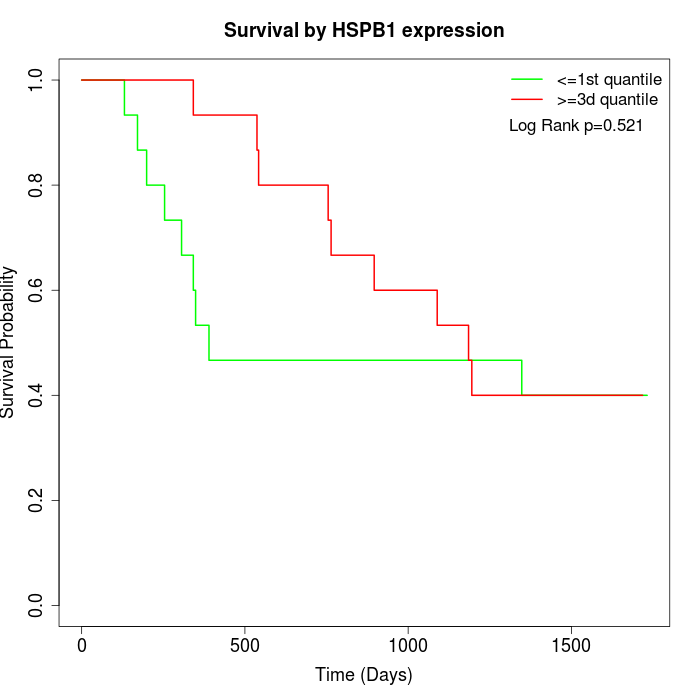
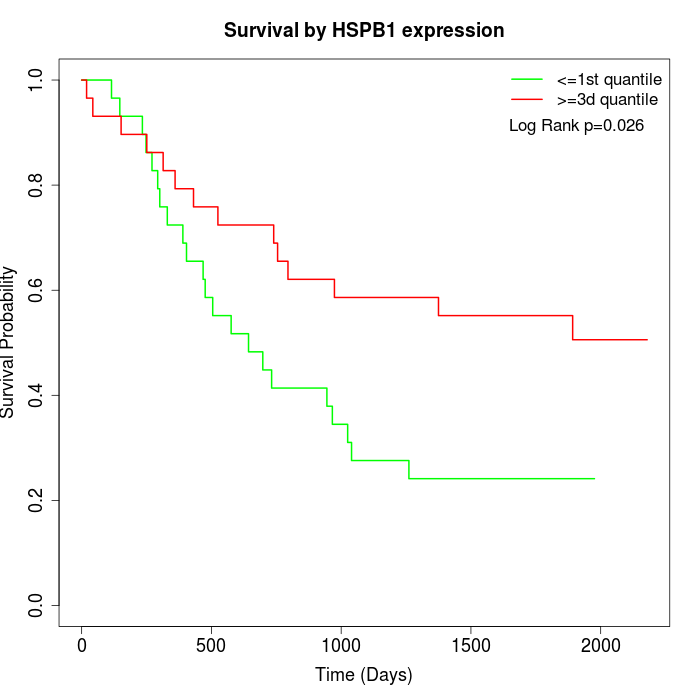
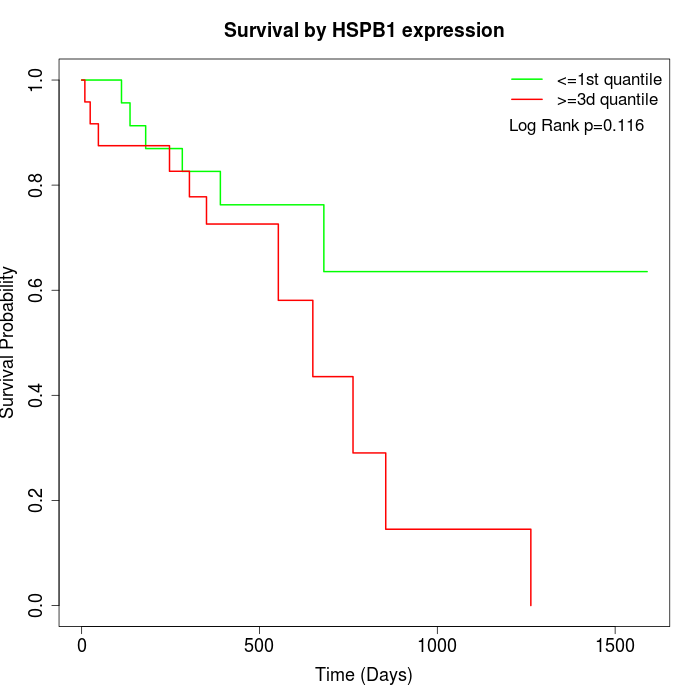
Survival by HSPB1 expression:
Note: Click image to view full size file.
Copy number change of HSPB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HSPB1 | 3315 | 14 | 1 | 15 | |
| GSE20123 | HSPB1 | 3315 | 14 | 1 | 15 | |
| GSE43470 | HSPB1 | 3315 | 3 | 2 | 38 | |
| GSE46452 | HSPB1 | 3315 | 11 | 0 | 48 | |
| GSE47630 | HSPB1 | 3315 | 7 | 2 | 31 | |
| GSE54993 | HSPB1 | 3315 | 2 | 7 | 61 | |
| GSE54994 | HSPB1 | 3315 | 14 | 3 | 36 | |
| GSE60625 | HSPB1 | 3315 | 0 | 0 | 11 | |
| GSE74703 | HSPB1 | 3315 | 3 | 1 | 32 | |
| GSE74704 | HSPB1 | 3315 | 8 | 1 | 11 | |
| TCGA | HSPB1 | 3315 | 45 | 5 | 46 |
Total number of gains: 121; Total number of losses: 23; Total Number of normals: 344.
Somatic mutations of HSPB1:
Generating mutation plots.
Highly correlated genes for HSPB1:
Showing top 20/456 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HSPB1 | RSBN1 | 0.677081 | 3 | 0 | 3 |
| HSPB1 | LYNX1 | 0.674258 | 4 | 0 | 4 |
| HSPB1 | HS3ST6 | 0.666366 | 3 | 0 | 3 |
| HSPB1 | VSIG10L | 0.650222 | 4 | 0 | 3 |
| HSPB1 | SLC9A9 | 0.646454 | 4 | 0 | 4 |
| HSPB1 | PRADC1 | 0.635708 | 5 | 0 | 4 |
| HSPB1 | KMT2D | 0.627393 | 4 | 0 | 4 |
| HSPB1 | CSTB | 0.624901 | 9 | 0 | 8 |
| HSPB1 | VPS25 | 0.622069 | 4 | 0 | 4 |
| HSPB1 | PITPNM3 | 0.620528 | 5 | 0 | 4 |
| HSPB1 | JUP | 0.619836 | 10 | 0 | 8 |
| HSPB1 | ANKRD18B | 0.619388 | 4 | 0 | 3 |
| HSPB1 | USH1G | 0.618844 | 4 | 0 | 4 |
| HSPB1 | MOB3C | 0.618185 | 4 | 0 | 4 |
| HSPB1 | SRSF4 | 0.617057 | 3 | 0 | 3 |
| HSPB1 | AGAP3 | 0.616374 | 3 | 0 | 3 |
| HSPB1 | MOCS2 | 0.616043 | 3 | 0 | 3 |
| HSPB1 | RHBDD1 | 0.614663 | 3 | 0 | 3 |
| HSPB1 | HCG22 | 0.608614 | 3 | 0 | 3 |
| HSPB1 | LGI3 | 0.608227 | 4 | 0 | 3 |
For details and further investigation, click here