| Full name: inhibitor of DNA binding 2 | Alias Symbol: GIG8|bHLHb26 | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 3398 | HGNC ID: HGNC:5361 | Ensembl Gene: ENSG00000115738 | OMIM ID: 600386 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ID2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells |
Expression of ID2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ID2 | 3398 | 201565_s_at | -0.4058 | 0.6068 | |
| GSE20347 | ID2 | 3398 | 201565_s_at | -0.1246 | 0.6917 | |
| GSE23400 | ID2 | 3398 | 201565_s_at | -0.2401 | 0.1953 | |
| GSE26886 | ID2 | 3398 | 201565_s_at | 0.3507 | 0.4285 | |
| GSE29001 | ID2 | 3398 | 201565_s_at | 0.1603 | 0.7134 | |
| GSE38129 | ID2 | 3398 | 201565_s_at | -0.3341 | 0.2571 | |
| GSE45670 | ID2 | 3398 | 201565_s_at | -0.4176 | 0.1418 | |
| GSE53622 | ID2 | 3398 | 103795 | -0.0534 | 0.7020 | |
| GSE53624 | ID2 | 3398 | 103795 | 0.0480 | 0.7083 | |
| GSE63941 | ID2 | 3398 | 201565_s_at | -3.3910 | 0.0185 | |
| GSE77861 | ID2 | 3398 | 201565_s_at | 0.7524 | 0.3164 | |
| GSE97050 | ID2 | 3398 | A_23_P143143 | -0.5215 | 0.4628 | |
| SRP007169 | ID2 | 3398 | RNAseq | -0.0063 | 0.9888 | |
| SRP008496 | ID2 | 3398 | RNAseq | 0.1315 | 0.7110 | |
| SRP064894 | ID2 | 3398 | RNAseq | -0.0426 | 0.8814 | |
| SRP133303 | ID2 | 3398 | RNAseq | 0.2105 | 0.3120 | |
| SRP159526 | ID2 | 3398 | RNAseq | 0.4501 | 0.3034 | |
| SRP193095 | ID2 | 3398 | RNAseq | 0.0522 | 0.8358 | |
| SRP219564 | ID2 | 3398 | RNAseq | -0.1521 | 0.7890 | |
| TCGA | ID2 | 3398 | RNAseq | -0.2294 | 0.0226 |
Upregulated datasets: 0; Downregulated datasets: 1.
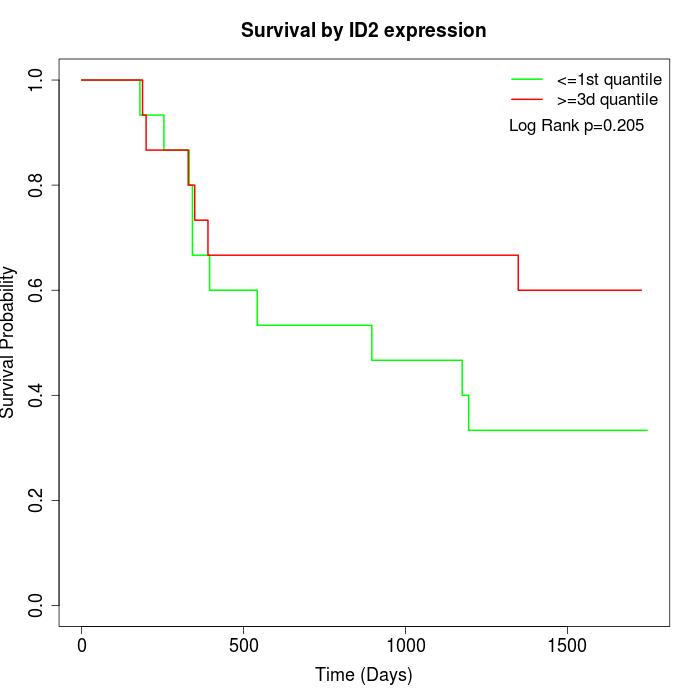
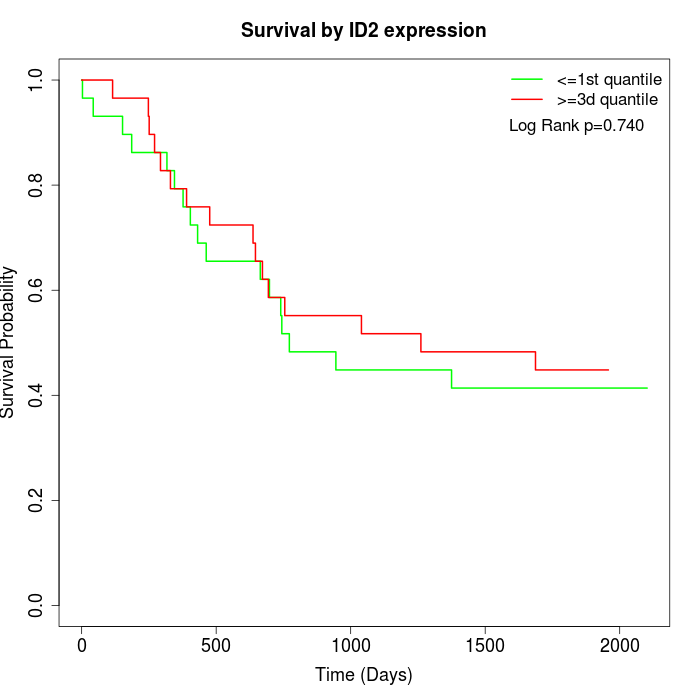
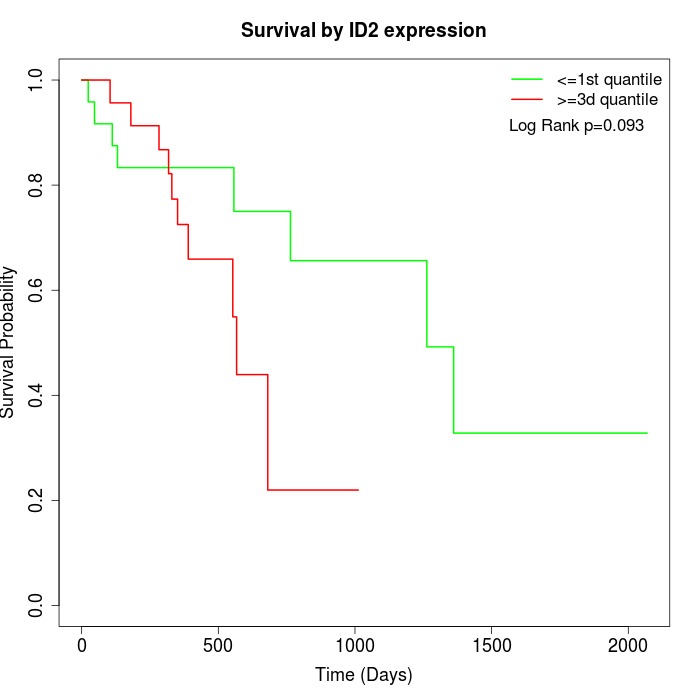
Survival by ID2 expression:
Note: Click image to view full size file.
Copy number change of ID2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ID2 | 3398 | 9 | 2 | 19 | |
| GSE20123 | ID2 | 3398 | 9 | 2 | 19 | |
| GSE43470 | ID2 | 3398 | 2 | 1 | 40 | |
| GSE46452 | ID2 | 3398 | 3 | 4 | 52 | |
| GSE47630 | ID2 | 3398 | 7 | 0 | 33 | |
| GSE54993 | ID2 | 3398 | 0 | 7 | 63 | |
| GSE54994 | ID2 | 3398 | 11 | 0 | 42 | |
| GSE60625 | ID2 | 3398 | 0 | 3 | 8 | |
| GSE74703 | ID2 | 3398 | 2 | 0 | 34 | |
| GSE74704 | ID2 | 3398 | 9 | 1 | 10 | |
| TCGA | ID2 | 3398 | 34 | 4 | 58 |
Total number of gains: 86; Total number of losses: 24; Total Number of normals: 378.
Somatic mutations of ID2:
Generating mutation plots.
Highly correlated genes for ID2:
Showing top 20/554 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ID2 | CHCHD10 | 0.781489 | 3 | 0 | 3 |
| ID2 | C11orf96 | 0.755892 | 3 | 0 | 3 |
| ID2 | CD99L2 | 0.744096 | 3 | 0 | 3 |
| ID2 | CCDC149 | 0.721137 | 4 | 0 | 4 |
| ID2 | DTX3 | 0.721002 | 3 | 0 | 3 |
| ID2 | C20orf194 | 0.719142 | 3 | 0 | 3 |
| ID2 | SLC25A25 | 0.70852 | 3 | 0 | 3 |
| ID2 | SYNC | 0.701368 | 4 | 0 | 4 |
| ID2 | BCL6 | 0.688173 | 3 | 0 | 3 |
| ID2 | NKTR | 0.682948 | 3 | 0 | 3 |
| ID2 | ABCD2 | 0.679667 | 3 | 0 | 3 |
| ID2 | CNN1 | 0.678856 | 4 | 0 | 4 |
| ID2 | MYEF2 | 0.675316 | 4 | 0 | 3 |
| ID2 | KANK2 | 0.669477 | 5 | 0 | 4 |
| ID2 | MCAM | 0.663193 | 5 | 0 | 4 |
| ID2 | HIPK3 | 0.662808 | 4 | 0 | 3 |
| ID2 | DDX42 | 0.661533 | 3 | 0 | 3 |
| ID2 | IL33 | 0.661125 | 5 | 0 | 4 |
| ID2 | FBN1 | 0.65202 | 5 | 0 | 4 |
| ID2 | FKBP7 | 0.648709 | 4 | 0 | 3 |
For details and further investigation, click here